Common questions in molecular biology: What are oligonucleotides used for?
A workhorse of molecular biology, oligonucleotides are relatively short single-stranded sequences of nucleotides, the monomeric subunits of both DNA and RNA. Oligonucleotides (often called oligomers or “oligos”) are invaluable in many molecular biology techniques, including PCR, DNA sequencing, labeled probes, plasmid construction, and genomic manipulations. Also used in microarrays, in situ hybridization, antisense analyses, and as drug carriers, oligos are now being tested in disease diagnosis and therapeutics, further advancing biological research and biomedicine.
The properties of complementarity and sequence specificity are key to the usefulness and versatility of oligonucleotides, as are their relative chemical stability, manipulability, and ease of labeling. Complementarity is the chemical recognition and hydrogen-bonding between specific nucleotide bases, which drives the formation of double-stranded molecules and maintains the integrity of DNA sequence. Complementary base pairs – adenine and thymine (DNA)/uracil (RNA), and guanine and cytosine – find each other during the process of annealing to form double-stranded nucleic acid molecules. The specific hydrogen bonding between complementary nucleotides dictates sequence specificity and preserves sequence integrity, making oligonucleotides valuable as probes and other molecular tools.
Uses of oligonucleotide primers
DNA polymerase, the enzyme of DNA replication, requires single-stranded oligos known as primers to initiate DNA synthesis. A primer, either of DNA or RNA, bound to a template sequence prompts the DNA polymerase to produce the complementary strand. DNA oligos are preferred due to their greater stability. DNA replication in vitro is performed for purposes of DNA sequencing, PCR, and labeled probe synthesis.
Oligonucleotide primer-based technologies can be used anytime nucleic acids are involved.
The use of polymerase chain reaction (PCR) revolutionized the study of gene expression and disease processes. To generate sufficient DNA material for study from a very small sample, PCR requires pairs of specific primers to rapidly copy a target DNA sequence. Oligonucleotide primers are also used for DNA Sequencing. Sequencing primers resemble those for PCR but vary slightly due to different polymerase reaction conditions. Plasmids, small circular DNA molecules widely used in recombinant DNA techniques, are often constructed with oligonucleotide primer insertions as a means of transfer engineered genetic information between cells. In short, oligonucleotide primer-based technologies can be used anytime nucleic acids are involved.
Oligonucleotide probes
Oligonucleotide probes are used for detecting RNA or DNA sequences of interest. Probes are designed to be complementary to the sequence of interest and are labeled with detectable molecules, such as fluorophores. Such probes are hybridized to immobilized target DNA or RNA to quantify the target sequence or localize it in situ. Adjusting the stringency of the hybridization conditions affects probe specificity, such that high stringency requires highly similar sequences and lower stringency decreases the required similarity.
Oligonucleotide probes are routinely used for screening gene libraries, in situ hybridization, gene expression analysis, and tissue microarrays.
Novel uses of oligonucleotides
Novel applications developed for oligonucleotides include aptamers and antisense oligonucleotides. Aptamers are designed to bind one or more specific targets for use in many of the same applications as antibodies. Aptamers are used as drugs and drug delivery systems, for identifying molecular markers of disease, and as molecular switches for various purposes, including in some biosensors (Li).
In a technique known as antisense therapy, antisense oligonucleotides (ASOs) target mRNA expression during disease processes through various mechanisms, including ribonuclease-mediated decay of pre-mRNA, direct steric blockage, and exon sequence modulation by binding the pre-mRNA at the splice site. Antisense therapy has proven effective against several diseases, including Duchenne muscular dystrophy and spinal muscular atrophy (Kim).
Oligonucleotides serve as DNA barcodes, used as insertions in various types of DNA libraries for identity and tracking purposes.
Oligonucleotides also serve as DNA barcodes by connecting a specific DNA sequence to the identity of a target molecule. Inspired by unique naturally-occurring sequences used to identify organisms in the wild, engineered DNA barcodes incorporate unique sequences into experimentally relevant DNA for identification purposes. Lab-derived DNA barcodes are used as insertions in various types of DNA libraries for identity and tracking purposes (Chen) since their unique design provides a reliable means of identifying sequences of interest.
Oligonucleotides for high-throughput gene expression analysis

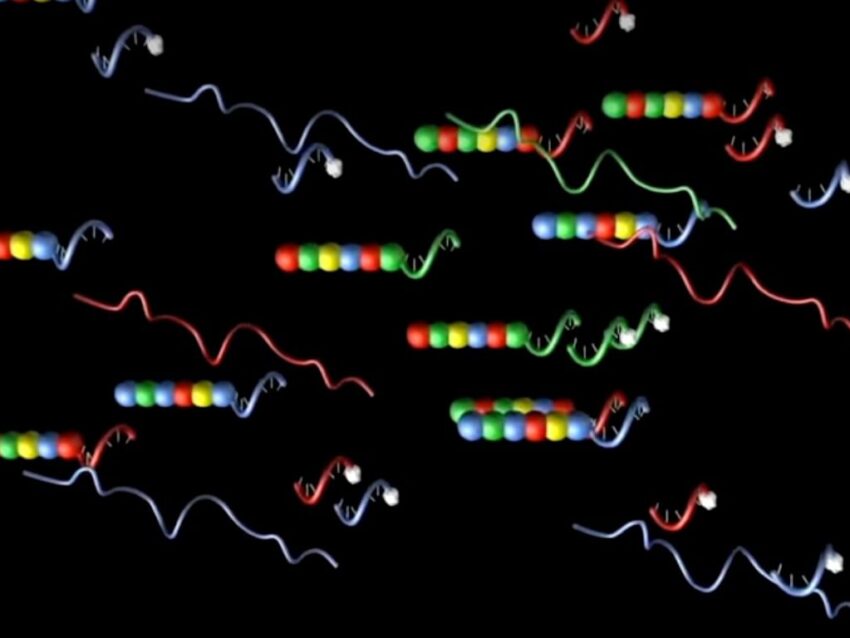
A variation on the concept of DNA barcodes uses oligonucleotides in molecular barcodes to tag mRNA transcripts in gene expression profiling studies of tissues. Pioneered by NanoString for use in the nCounter® Pro Analysis System, this technology uses both DNA and RNA oligos to form multi-part probes for identifying and quantifying transcripts expressed in a tissue. Three specifically-designed unique oligonucleotides (two 50mer DNA oligos and an RNA molecular barcode complementary to one of the 50mers) constitute the heart of each nCounter probe. Individual segments of the RNA oligo are labeled with specific fluorophores arranged in specific linear orders to create unique codes for detecting and quantifying each transcript of interest (Geiss).
The elegance of nCounter Pro’s fluorescent molecular barcode probes is the latest example demonstrating the priceless role oligonucleotides play in modern biotechnology.
References
Chen BR, Hale DC, Ciolek PJ, Runge KW. Generation and analysis of a barcode-tagged insertion mutant library in the fission yeast Schizosaccharomyces pombe. BMC Genomics. 2012;13:161. Published 2012 May 3. doi:10.1186/1471-2164-13-161
Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008 Mar;26(3):317-25. doi: 10.1038/nbt1385. Epub 2008 Feb 17. PMID: 18278033.

Related Content