
Make the Leap to Single-Cell Spatial and Unlock Unprecedented Biology with 6000 Genes
Are you developing a single-cell atlas, identifying functional cell states, defining ligand-receptor interactome, characterizing tissue microenvironment phenotypes or doing biomarker discovery? If so, the CosMx™ 6K Discovery panel enables you to spatially analyze virtually the entire reactome at the single cell level and achieve breakthrough results.
The CosMx™ SMI and decoder probes are not offered and/or delivered to the Federal Republic of Germany for use in the Federal Republic of Germany for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof in a method used in fluorescence in situ hybridization for detecting a plurality of analytes in a sample without the consent of the President and Fellows of Harvard College (Harvard Corporation) as owner of the German part of EP 2 794 928 B1. The use for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof is prohibited without the consent of the President and Fellows of Harvard College (Harvard Corporation).
Applications
Semi-supervised cell typing using Allen Human M1 Atlas
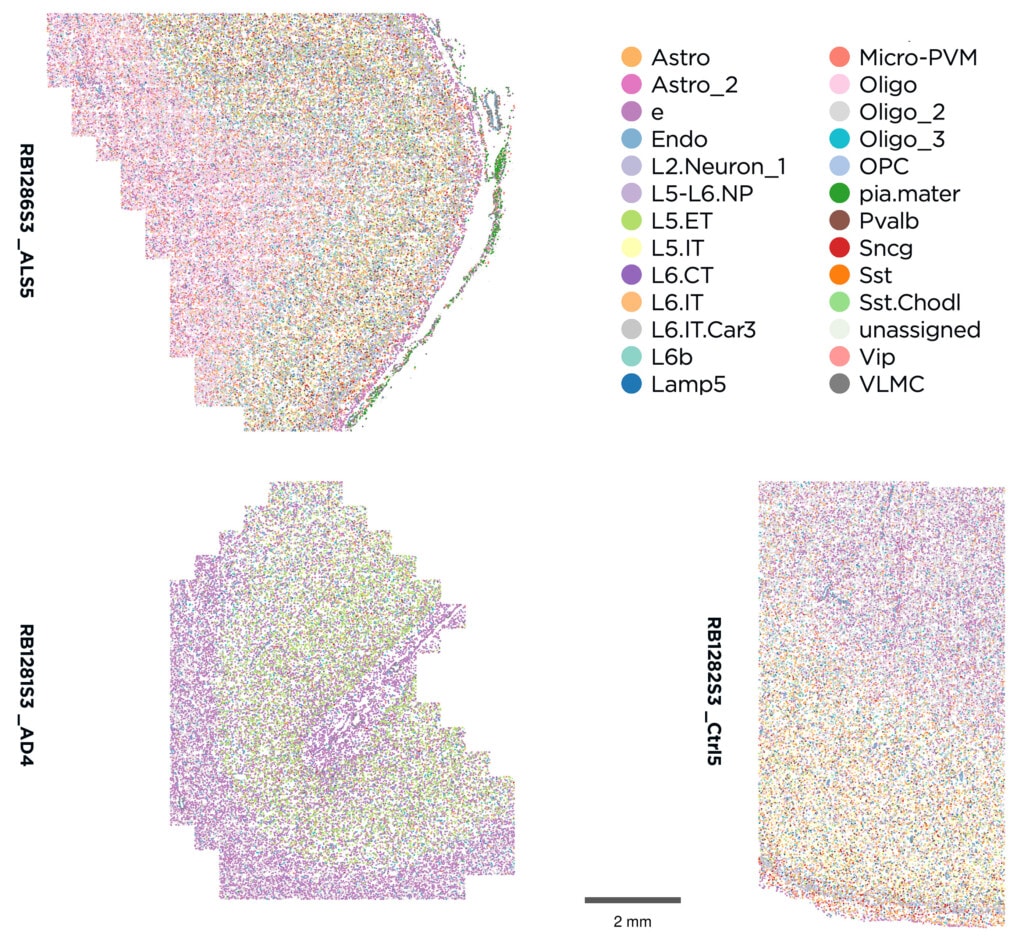
nb_clus #1 (motorClus20_custom_qc_2)
Novel clusters: Astro_2, L2.Neuron_1, Oligo_2, Oligo_3, e, pia.mater
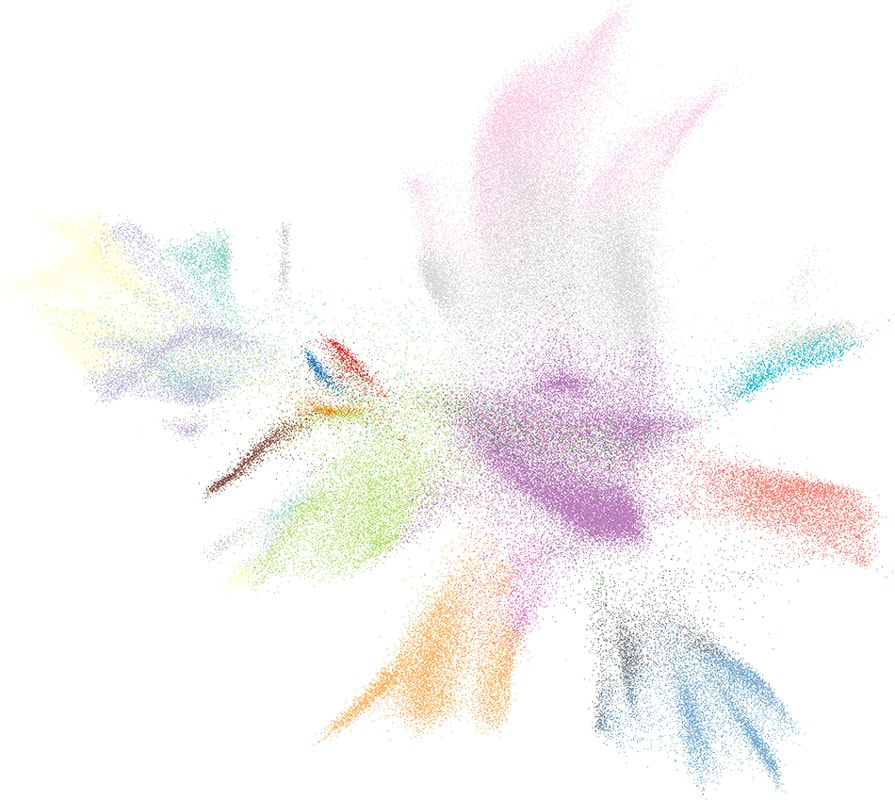
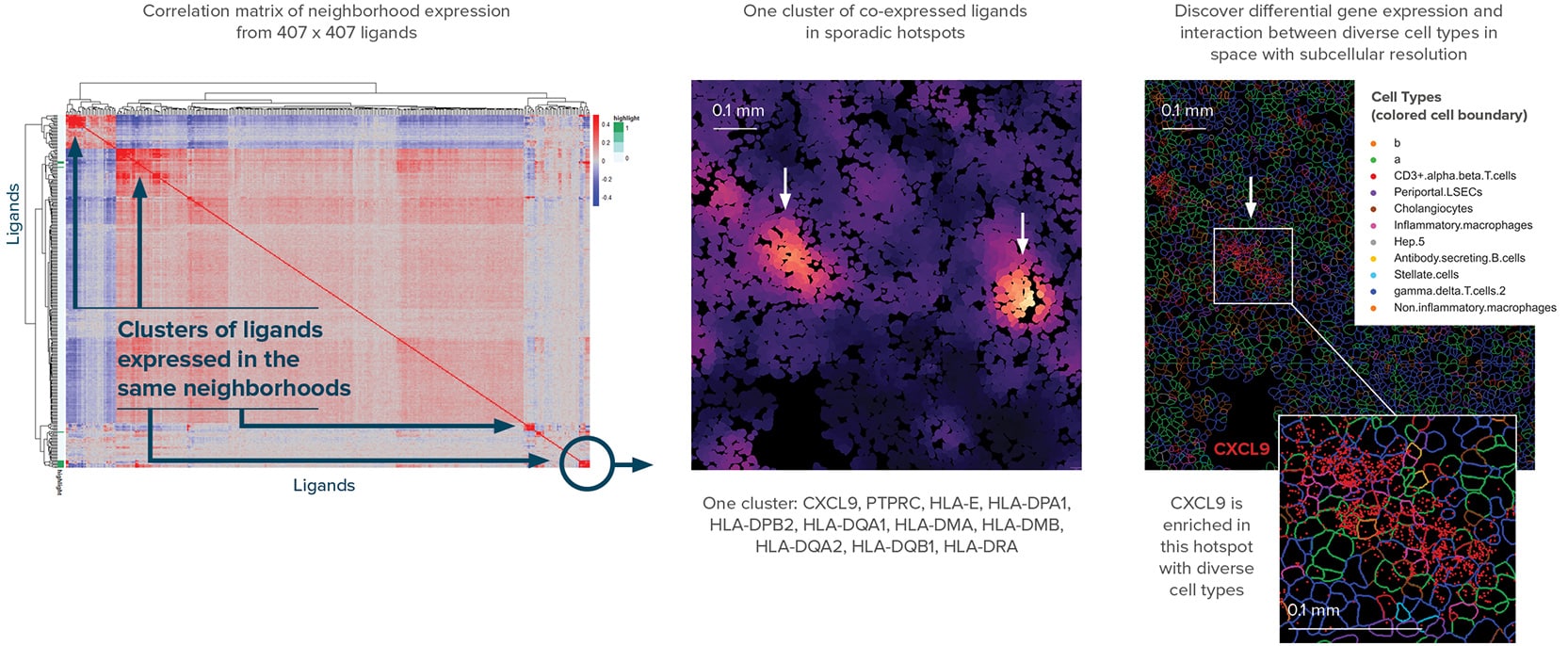
Discovery of spatially co-expressed ligands in diverse types of cells in FFPE liver cancer
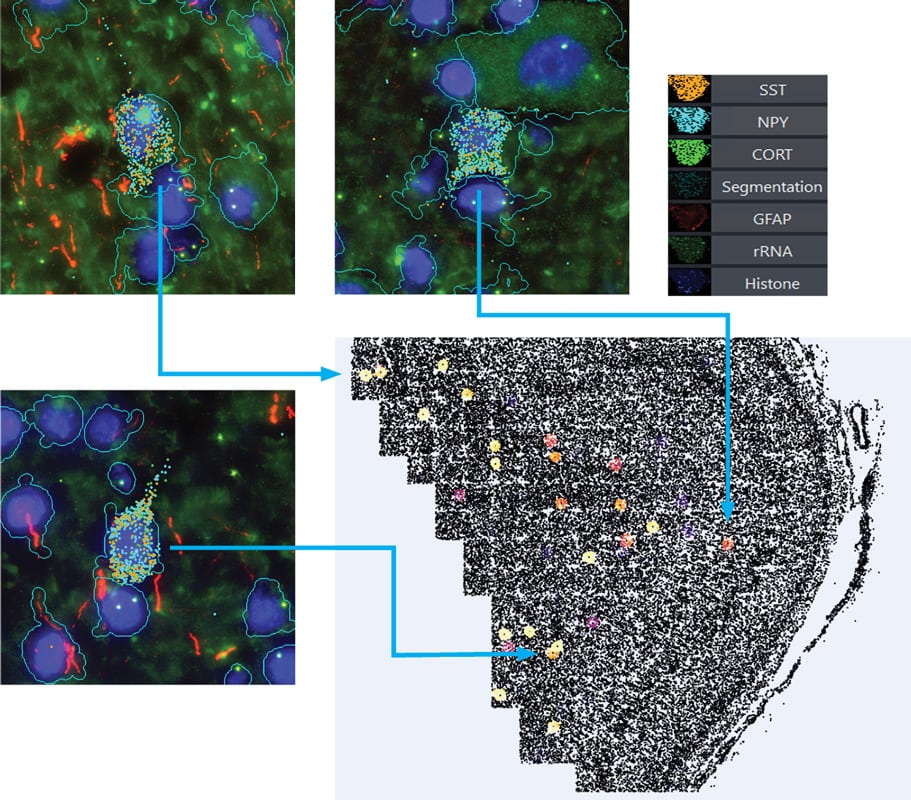
Ligands Co-localization Identify Rare Cell Type in Human Brain: Sst Chodl neurons
Unsupervised clustering is performed on the correlation matrix of ligands’ neighborhood expression profiles to identify clusters of co-localized ligands. One ligand cluster consists of 3 ligands, SST, NPY, CORT, and shows strong correlation in their neighborhood expression profiles. The co-expression of those 3 ligands identify a rare cell type, Sst Chodl interneurons, in the dataset.
Plex does matter.
The utilization of the CosMx 6K Discovery Panel enables a more comprehensive analysis of Differential Expression and significantly enhances the potential for biomarker discovery. As shown across the two volcano plots, the CosMx 6K Discovery Panel identifies a larger number of statistically significant and biologically meaningful genes associated with Kidney cancer as compared to the CosMx Universal Cell Characterization (1K) Panel.
Why use CosMx 6K Discovery Panel over scRNA-seq?
Request a Quote
Contact our helpful experts and we’ll be in touch soon.
Research
AACR 2024: Reveal spatial signatures of TME and oncogenic pathways using 6K plex single-cell spatial molecular imaging on FFPE tissue
Ultra High-Plex Single-Cell Spatial Multiomic Imaging of Human Brain Sections – AGBT 2024
A Universal 6,000-plex RNA Panel to Construct Comprehensive Single-Cell Spatial Atlases Across Multiple FFPE Human Tissues – AGBT 2024
Path to the holy grail of spatial biology: Spatial single-cell whole transcriptomes using 6000-plex spatial molecular imaging on FFPE tissue – AACR 2023
High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging
Resolving the spatial distribution of RNA and protein in tissues at subcellular resolution is a challenge in the field of spatial biology. We describe spatial molecular imaging, a system that measures RNAs and proteins in intact biological samples at subcellular resolution by performing multiple cycles of nucleic acid hybridization of fluorescent molecular barcodes.




