GeoMx® Human Whole Transcriptome Atlas
Helping Your Research
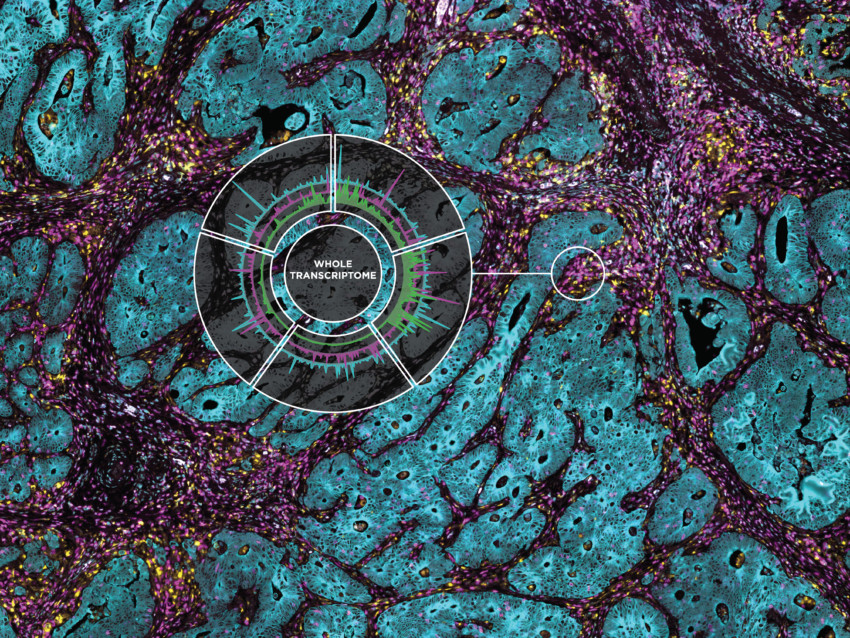
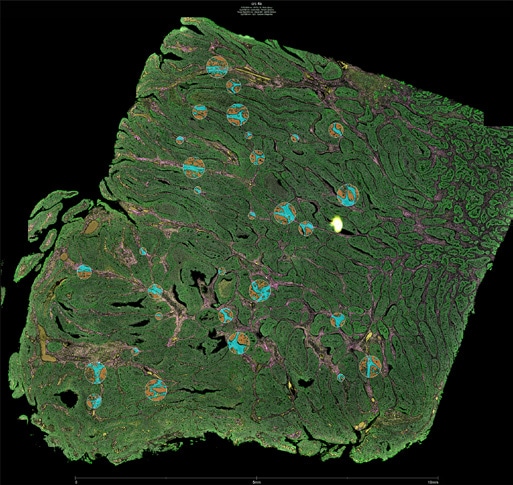
Full Transcriptome Spatial RNA Analysis
The GeoMx Human Whole Transcriptome Atlas (GeoMx Hu WTA) is designed for comprehensive profiling of spatial biology. With full coverage of protein coding genes, GeoMx Hu WTA delivers spatial analysis of any target in any tissue in the biological regions that matter most.


Publications
Opposing immune and genetic mechanisms shape oncogenic programs in synovial sarcoma
Synovial sarcoma (SyS) is an aggressive neoplasm driven by the SS18-SSX fusion, and is characterized by low T cell infiltration. Here, we studied the cancer-immune interplay in SyS using an integrative approach that combines single-cell RNA sequencing (scRNA-seq), spatial profiling and genetic and pharmacological perturbations.
The spatial landscape of lung pathology during COVID-19 progression
Recent studies have provided insights into the pathology and immune response to coronavirus disease 2019 (COVID-19)1–8. However, thorough interrogation of the interplay between infected cells and the immune system at sites of infection is lacking.
Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection
The relationship of SARS-CoV-2 pulmonary infection and severity of disease is not fully understood. Here we show analysis of autopsy specimens from 24 patients who succumbed to SARS-CoV-2 infection using a combination of different RNA and protein analytical platforms to characterize inter-patient and intra-patient heterogeneity of pulmonary virus infection.
Related Resources

Request a Quote
Contact our helpful experts and we’ll be in touch soon.