The Challenges of Reverse Transcription (RT)
For years, scientists have relied on platforms that use reverse transcriptase to convert RNA to cDNA in order to perform gene expression studies. However, the cDNA conversion and amplification steps can introduce variability into the data. Additionally, other steps in the workflow like sample prep and data analysis can introduce variability. The scientific community is now recognizing these challenges, and publications on the limitations of RT-based platforms are emerging across the qPCR, microarray and NGS spaces.
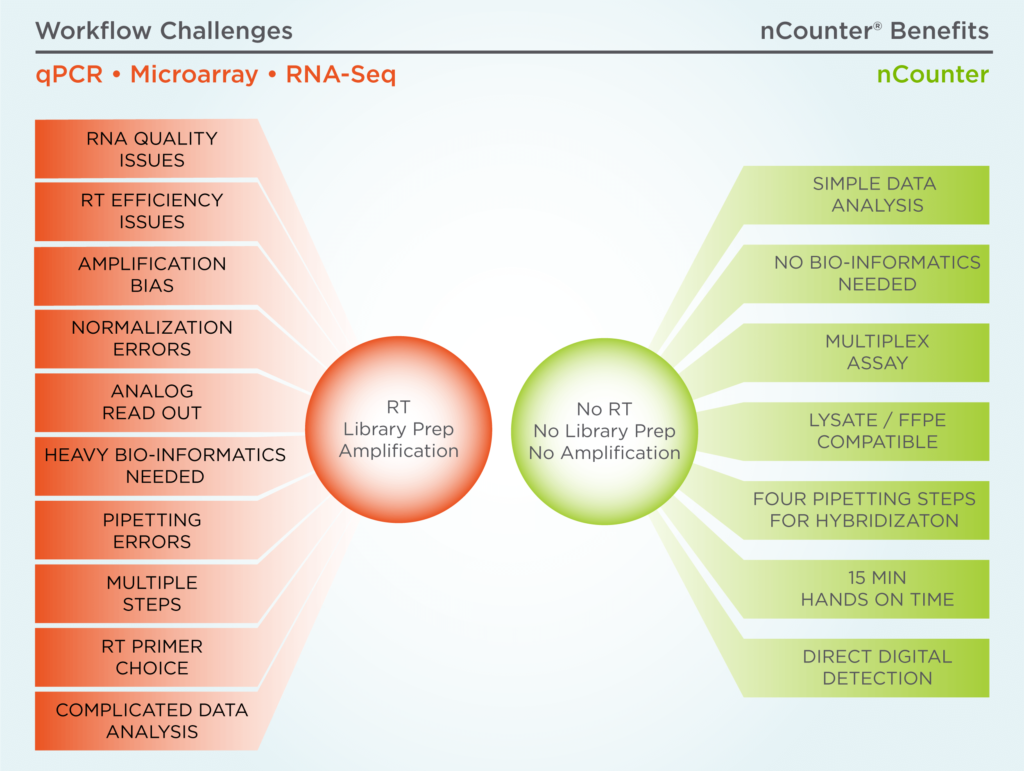
The Benefits of Counting RNA Directly
What if you could bypass a cDNA conversion step and count RNA molecules directly? The nCounter® platform offers direct RNA detection for robust, reproducible performance and unbiased transcript quantitation.
Interested in learning more? Your journey begins here.
Publications
Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples
Adaptation of a RAS pathway activation signature from FF to FFPE tissues in colorectal cancer
Apparent bias toward long gene misregulation in MeCP2 syndromes disappears after controlling for baseline variations
Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples
Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries
Despite the ever-increasing output of Illumina sequencing data, loci with extreme base compositions are often under-represented or absent. To evaluate sources of base-composition bias, we traced genomic sequences ranging from 6% to 90% GC through the process by quantitative PCR.
Talking the talk, but not walking the walk: RT-qPCR as a paradigm for the lack of reproducibility in molecular research
Poorly executed and inadequately reported molecular measurement methods are amongst the causes underlying the lack of reproducibility of much biomedical research. Although several high impact factor journals have acknowledged their past failure to scrutinise adequately the technical soundness of manuscripts, there is a perplexing reluctance to implement basic corrective measures.
The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments
Background: Currently, a lack of consensus exists on how best to perform and interpret quantitative real-time PCR (qPCR) experiments. The problem is exacerbated by a lack of sufficient experimental detail in many publications, which impedes a reader’s ability to evaluate critically the quality of the results presented or to repeat the experiments.
Variability of the reverse transcription step: practical implications
Background: The reverse transcription (RT) of RNA to cDNA is a necessary first step for numerous research and molecular diagnostic applications. Although RT efficiency is known to be variable, little attention has been paid to the practical implications of that variability.
Sources of error in the retracted scientific literature
Retraction of flawed articles is an important mechanism for correction of the scientific literature. We recently reported that the majority of retractions are associated with scientific misconduct.
Critical appraisal of quantitative PCR results in colorectal cancer research: can we rely on published qPCR results?
The use of real-time quantitative polymerase chain reaction (qPCR) in cancer research has become ubiquitous. The relative simplicity of qPCR experiments, which deliver fast and cost-effective results, means that each year an increasing number of papers utilizing this technique are being published.
A Comparison of RNA-Seq Results from Paired Formalin-Fixed Paraffin-Embedded and Fresh-Frozen Glioblastoma Tissue Samples
The molecular classification of glioblastoma (GBM) based on gene expression might better explain outcome and response to treatment than clinical factors. Whole transcriptome sequencing using next-generation sequencing platforms is rapidly becoming accepted as a tool for measuring gene expression for both research and clinical use.
Library construction for next-generation sequencing: Overviews and challenges
High-throughput sequencing, also known as next-generation sequencing (NGS), has revolutionized genomic research. In recent years, NGS technology has steadily improved, with costs dropping and the number and range of sequencing applications increasing exponentially.
Standards and Guidelines for Validating Next-Generation Sequencing Bioinformatics Pipelines: A Joint Recommendation of the Association for Molecular Pathology and the College of American Pathologists
Bioinformatics pipelines are an integral component of next-generation sequencing (NGS). Processing raw sequence data to detect genomic alterations has significant impact on disease management and patient care.
Amplification of nonspecific products in quantitative polymerase chain reactions (qPCR)
Quantitative PCR allows the precise measurement of DNA concentrations and is generally considered to be straightforward and trouble free. However, a survey with 93 validated assays for genes in the Wnt-pathway showed that the amplification of nonspecific products occurs frequently and is unrelated to Cq or PCR efficiency values.