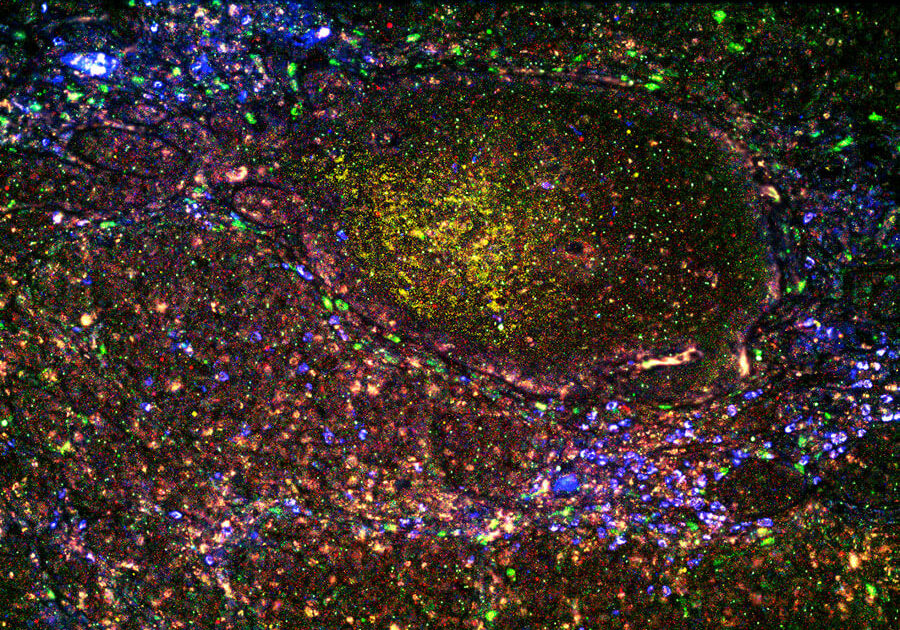
Spatial Genomics
The field of spatial genomics is entering its teenage years. Since its first description in 2009, scientists have used the phrase spatial genomics, spatially resolved transcriptomics, spatial biology, spatial sequencing, and spatial profiling to describe the technique. Regardless of the term used, spatial genomics is a promising and exciting technology. For instance, Nature Methods chose spatially resolved transcriptomics as the method of the year in 2020.
What is spatial genomics?
Spatial genomics is the study of the content and expression of the genome within its native spatial context.
Spatial genomics refers to assessing a cell’s genomics while maintaining biological context. One of the central tenets of molecular biology is that form dictates function; the biological context is critical for understanding the functionality and importance of a cell’s genomics. Therefore, spatial genomics provides a sweet spot for molecular biologists who want to understand the biological importance of transcriptional dynamics in context.
Spatial genomics is the novel combination of standard molecular biology techniques, including standard immunofluorescence techniques, digital optical barcoding technology, next-generation sequencing, and in situ hybridization, such as the latest NanoString single-cell imaging technology. These techniques allow scientists to examine transcripts within their spatial context. Structural and positional context is important for understanding molecular function. Therefore, spatially resolved transcriptomics can reveal critical insight into cellular function.
With this information, one can better understand cellular behavior, social context, and transcriptional dynamics during cellular and/or molecular processes. In addition, researchers can examine how transcriptional dynamics vary within a tissue or between diseased and non-diseased tissues. Spatial genomics techniques are not limited to the tissue or cellular level. Users may also examine subcellular transcriptional dynamics. For instance, examining mRNA transcript distribution within different functional regions of a cell.
Three Platforms. Unlimited Potential.
Whether you’re characterizing with standard gene expression or exploring with novel spatial biology approaches, NanoString’s platforms and integrated analytics provide robust and reproducible technologies to bring progress to your work.
What is spatial gene expression?
Spatial gene expression is a method for studying transcriptional dynamics in their three-dimensional cellular context.
Biological systems exist in three-dimensional space. Notably, the three-dimensional context is critical for proper functioning. Many molecular processes require cells or molecules to be in a precise orientation relative to one other. For example, the binding of ligands to an enzyme or the position of notch and delta on adjacent cells to activate notch signaling in the target cell. Therefore, attention to spatial context is important for understanding the function of transcriptional dynamics during molecular processes.
Spatial gene expression is similar and sometimes used interchangeably with spatial genomics (described above). Like spatial genomics, spatial gene expression relies on well-known molecular biology techniques that together allow the user to examine transcriptional dynamics in a spatial context.
Spatial gene expression within an organ
Although the two terms are sometimes used interchangeably, the choice of term used reflects the user’s question. For example, one may be interested in gene expression dynamics in a variety of samples from one region of an organ. This approach would give a more general look at typically expressed genes in a specific healthy organ. One could say they examined the spatially resolved genomics in a particular region of interest in an organ and found some general patterns.
For example, some scientists are working to create a spatial organ atlas. The spatial organ atlas contains information on the general expression of genes within different functional locations in healthy organs. Researchers sequenced these functional regions in a variety of healthy samples in order to characterize gene expression dynamics. Using this as a reference, others can compare diseased tissues to standard expression and understand how genomics change.
If we understand how transcriptional dynamics change over time as a disease develops, it may also open the door for identifying new treatments for disease. It may also be possible to identify when a disease is developing. With this knowledge in hand, steps could be taken to prevent disease.
Spatial gene expression within a tumor
Spatial gene expression may be used as a more precise term. Rather than getting general information about gene expression in a particular region, a user may be comparing or manipulating the cells or tissues in question. A researcher may examine how gene expression differs in two parts of a tumor.
In a real-world example, a research group compared gene expression between low- and high-grade tumors in colorectal cancer. Another possible spatial gene expression question is to compare gene expression dynamics before and after treatment with some clinically relevant compound.
Why is spatial genomics important?
Spatial genomics is important because it provides detailed transcriptional information in a specific cellular context.
Genomics is a subfield of molecular biology that examines the structure, function, evolution, and mapping of genomes. Genomes refer to an organism’s DNA. Gene expression is the process of making an mRNA copy of the DNA (transcription) and translating that mRNA into a protein. These proteins (or, in some cases, RNA species) cause a biological change.
Gene expression varies between cells and tissue types. For instance, variance in gene expression is why we have different cell types, and yet all cells contain the same DNA. Since genomics translates to a biological change in the body, and the structure-function relationship within three-dimensional space within a cell are critical for biology, the combination of the two perspectives is of critical importance for fully understanding the molecular biology of cells.
Spatial genomics adds an additional layer of complexity to our understanding of cell biology, gene expression, and genomics. The study of this additional layer of complexity may revolutionize cell and molecular biology. Ultimately, this enhanced understanding of cell biology and transcriptional dynamics may lead to new strategies for the prevention and treatment of diseases.
A cell’s context is crucial for proper functioning. Assessment of gene expression in context gives scientists a more complete view of the molecular biology of cells.
Let’s make an analogy.
Say you want to characterize a particular landscape from the air. Bulk sequencing is like a blurry image of a large area. You can determine basic features but lose fine details. Single-cell sequencing and single-cell imaging are like having a high-resolution image of one part of it. Let’s say it’s an image of a tree. Now you have very specific information about a tree in your landscape. You can identify its shape, color, and if it has needles or leaves. However, you don’t know what the tree is near, if it’s the only tree or the only tree of that kind.
Spatial genomics is like taking a high-resolution image of the landscape. You can see details, like trees, plants, roads, and buildings. With single-cell imaging and sequencing, you can also see the details of the tree and determine what kind of tree it is. At the same time, you can see the context. Where the different landscape features are in relationship to one another. With the detailed context in hand, you can then make better inferences about function. For example, if the landscape you are viewing is a tree farm, home in the woods, or park in a city.
What is spatial sequencing?
Spatial sequencing is the process of characterizing all transcripts found in a biologically meaningful area or region.
Using spatial sequencing, a user can map variation in gene expression in different regions across a tissue or organ type. For example, these regions could include different functional compartments within a single organ, tumors in the same or different patients, diseased or healthy regions of the same tissue, or the interface between different tissue or cell types.
Since spatial orientation is a critical part of biological function, attention to the spatial distributions of transcripts is necessary for a holistic understanding of cellular function and biology. Understanding the spatial context for transcripts is critical for interpreting the biological meaning. Applications of spatial sequencing include cancer biology, pathology, and developmental biology.
How does spatial sequencing work?
Spatial sequencing works by combining two molecular biology techniques, microscopy and sequencing.
Spatial sequencing relies on the fusion of two common molecular biology techniques, immunofluorescence microscopy and next-generation sequencing. In addition, depending on the research question, spatial sequencing can be leveraged at the tissue, single cell, or subcellular levels.
Samples do not require special handling because standard molecular biology techniques are used. Instead, researchers can use standard preservation techniques like formalin-fixed paraffin-embedded or fresh-frozen samples. If the user wants to examine protein, only one to five large cells or ten to twenty small cells are necessary as starting materials. More cells are needed for RNA analysis, five to twenty large or fifty to two hundred small cells.
Once the samples are collected, the user labels either mRNA or protein with fluorescently labeled RNA probes or antibodies, respectively. After labeling, the samples are loaded onto a microscope, and the user chooses a region of interest. Once this region is identified, the user shines ultraviolet light onto the sample. The ultraviolet light causes the release of the molecular tags from the RNA probe or antibody. These tags are then collected, processed, and sequenced using next-generation sequencing to determine the transcripts present.
Spatial Genomics Applications
Applications of spatial genomics span molecular biology and include a variety of subdisciplines.
Context and spatial orientation is critical for understanding cellular and molecular function. Spatial genomics allows unprecedented insight into the biological context of transcriptional dynamics. As such, applications of spatial genomics span a wide range of fields under the umbrella of molecular biology.
Cancer Biology
Using spatial genomics, users can compare gene expression between low- and high-grade tumors, different regions within a single tumor, or tumors from different patients. For example, one research group found that expression of a specific gene called IDO1 varied between high and low-grade tumor regions in patients with colorectal cancer. Additionally, given the high specificity afforded using sophisticated microscopy, users can also examine the interface between a tumor and the immune system. For instance, Pelka et al. (2021) examined the transcriptional dynamics between tumor and immune cells in colorectal cancer.
Pathology
To understand disease progression, scientists must also understand normal transcriptional dynamics in healthy tissue. To that end, one can create a library or atlas of transcriptional dynamics not only in healthy tissue but in functionally different regions of a tissue or organ. For instance, one may wish to understand the transcriptional dynamics within the islets of Langerhans. The islets of Langerhans are important in the pancreas’ endocrine function. Therefore, it may be informative to examine transcriptional dynamics in the islets of Langerhans in individuals who are non-diabetic, pre-diabetic, and diabetic. This may provide critical insights into transcriptional dynamics underlying disease progression, potentially revealing critical biomarkers and/or clinical targets.
Developmental Biology
The process of a fertilized egg or zygote developing into a fully formed fetus is a complicated process. Many developmental processes rely on precise spatial orientation for various structures to develop properly. For example, patterning of certain structures, such as the limb, are dictated through differential exposure to proteins such as Sonic Hedgehog. Since Sonic Hedgehog is expressed in a gradient, areas of developing tissue exposed to high versus low levels form different structures. Therefore, one could examine transcriptional dynamics in different regions during pattern formation.
Other Spatial Genomics FAQs
Why is spatial genomics important?
Spatial genomics are important because the human body is a complex three-dimensional structure. That structure is an important part of our molecular physiology. To fully understand genomics requires that we examine it in a spatially resolved manner. Learn more »
What is spatial genomics & transcriptomics?
Spatial genomics includes genomic (total DNA and RNA), transcriptomic (RNA transcripts) and epigenomic (molecular compounds) profiling methods carried out in intact tissue with positional context. Spatial genomics enables molecularly connecting a particular genotype to its phenotype. Learn more »
What techniques are used in spatial genomics & transcriptomics?
Spatial genomics and spatial transcriptomics are naming variations for a technique whereby users can garner gene expression information in a spatial context. Using this technique, scientists can use next-generation sequencing to identify transcriptomics within a specific tissue or cellular context. Learn more »
Select Publications
Best Practices for Spatial Profiling for Breast Cancer Research with the GeoMx® Digital Spatial Profiler
Breast cancer is a heterogenous disease with variability in tumor cells and in the surrounding tumor microenvironment (TME). Understanding the molecular diversity in breast cancer is critical for improving prediction of therapeutic response and prognostication.
Digital quantitative assessment of PD-L1 using digital spatial profiling
The assessment of programmed death 1 ligand 1 (PD-L1) expression by Immunohistochemistry (IHC) is the US Food and Drug Administration (FDA)-approved predictive marker to select responders to checkpoint blockade anti-PD-1/PD-L1 axis immunotherapies. Different PD-L1 immunohistochemistry (IHC) assays use different antibodies and different scoring methods in tumor cells and immune cells.
Multiplex digital spatial profiling of proteins and RNA in fixed tissue
Digital Spatial Profiling (DSP) is a method for highly multiplex spatial profiling of proteins or RNAs suitable for use on formalin-fixed, paraffin-embedded (FFPE) samples. The approach relies on (1) multiplexed readout of proteins or RNAs using oligonucleotide tags; (2) oligonucleotide tags attached to affinity reagents (antibodies or RNA probes) through a photocleavable (PC) linker; and (3) photocleaving light projected onto the tissue sample to release PC oligonucleotides in any spatial pattern across a region of interest (ROI) covering 1 to ~5,000 cells.
Spatially organized multicellular immune hubs in human colorectal cancer
Immune responses to cancer are highly variable, with mismatch repair-deficient (MMRd) tumors exhibiting more anti-tumor immunity than mismatch repair-proficient (MMRp) tumors. To understand the rules governing these varied responses, we transcriptionally profiled 371,223 cells from colorectal tumors and adjacent normal tissues of 28 MMRp and 34 MMRd individuals.
A user’s perspective on GeoMx digital spatial profiling
Characterization of spatial protein expression for multiple targets from a single tissue is difficult to perform, especially due to the limitations of multiplex immunohistochemistry and tissue heterogeneity. Therefore, a new technology is required that permits detailed and simultaneous expression profiling of proteins within a defined region of interest (ROI).
Keep In Touch to Learn More