CosMx SMI Mouse Brain FFPE Dataset

Using the CosMx™ Spatial Molecular Imager (SMI), we characterized mouse brain FFPE tissue across two coronal sections, one covering a full hemisphere and the other spanning the hippocampal formation and cortex bilaterally. In this dataset, over 1500 transcripts were detected per cell with 800+ unique genes, highlighting the sensitivity of the platform and assay. The complete dataset consists of over 90,000 single cells and ~225 million transcripts to build a detailed atlas of the mouse brain. This high quality, single-cell tissue analysis provides a rich dataset to discover novel biology within the brain.
The CosMx™ SMI and decoder probes are not offered and/or delivered to the Federal Republic of Germany for use in the Federal Republic of Germany for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof in a method used in fluorescence in situ hybridization for detecting a plurality of analytes in a sample without the consent of the President and Fellows of Harvard College (Harvard Corporation) as owner of the German part of EP 2 794 928 B1. The use for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof is prohibited without the consent of the President and Fellows of Harvard College (Harvard Corporation).
How it Works
This dataset was generated using the CosMx Mouse Neuroscience RNA panel. This panel is fully validated and designed to provide robust cell typing, cell-cell interaction analysis and more in mouse brain and other neuronal tissues.
Data Files
Discover and Map Neuronal Cell Types
The CosMx Mouse Neuroscience panel delivers expression mapping of 1,000 target genes in a morphologically intact tissue. This fully validated and optimized assay is enabled to identify more than 40 distinct cell types in the mouse brain with curated content to provide information of cell states and interactions across neuroscience research areas including neuropathology and inflammation. Additionally, CosMx SMI utilizes RNA, protein and nuclear information to provide accurate defination of various cell shapes within the brain.

Unraveling the Complexity of Neurobiology
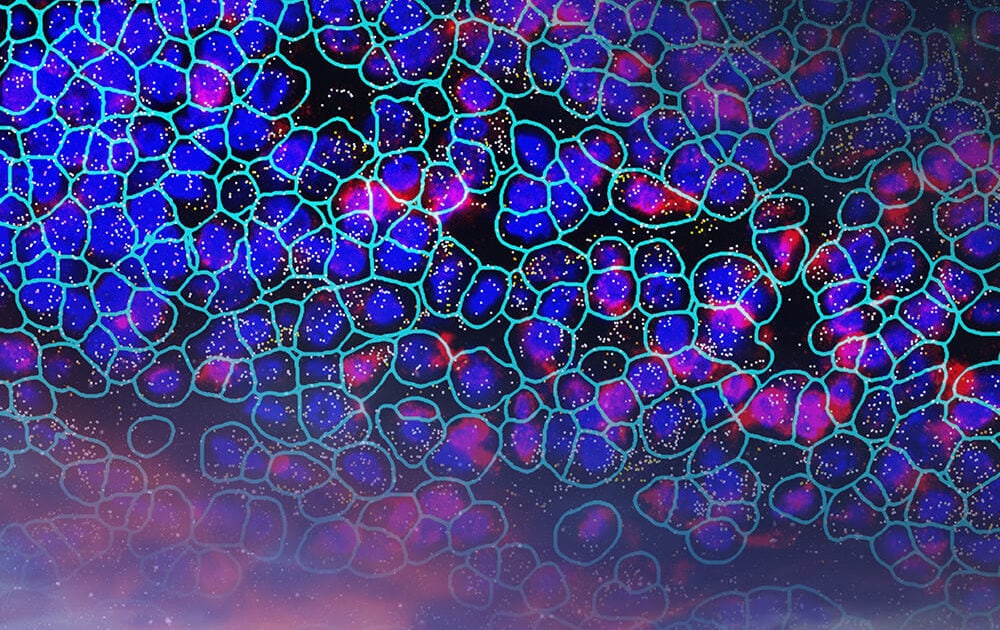
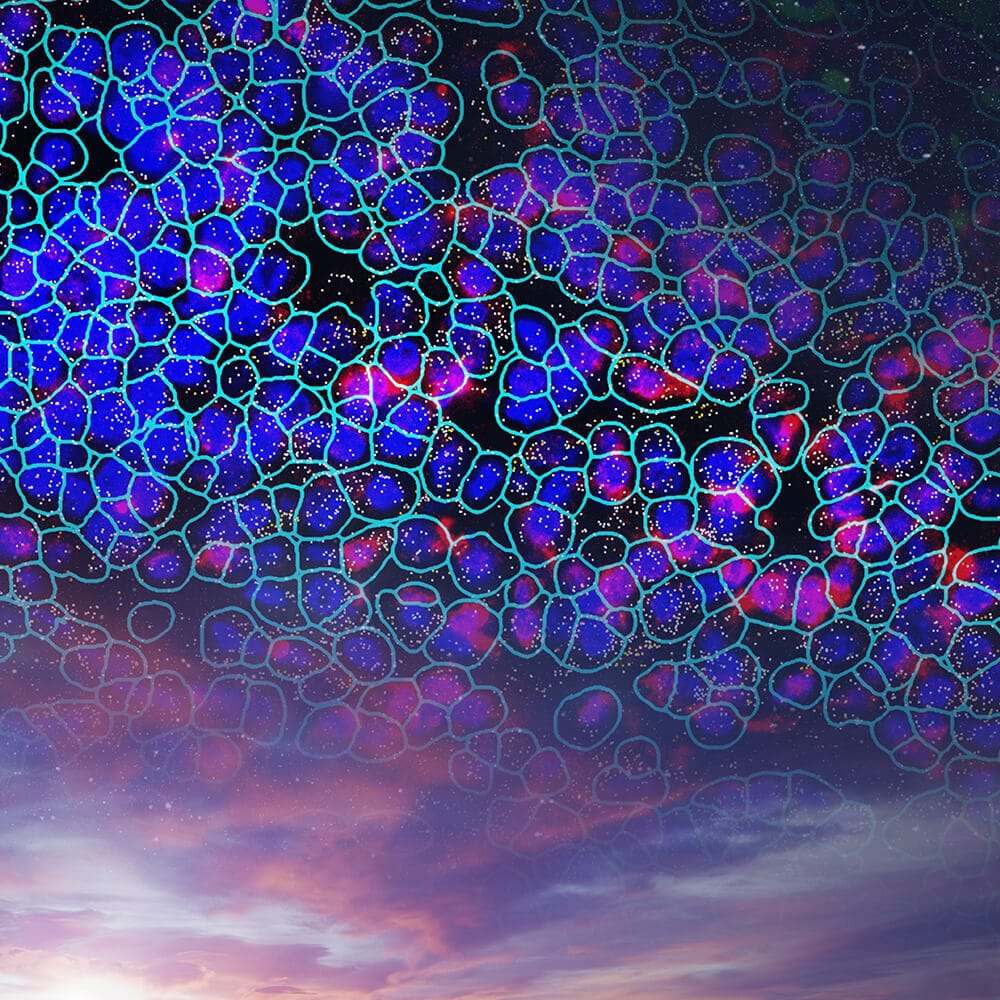
Emerging evidence supports a role for oligodendrocytes and oligodendrocyte precursor cells (OLPCs) in metabolic homeostasis within the hypothalamic arcuate nucleus. The CosMx Mouse Neuroscience panel enables researchers to unravel the complexities of this biological process. Spatially resolved data from within this CosMx SMI dataset captures the dynamic interaction between these oligodendrocytes and NPY-positive inhibitory interneurons (INHINs), neuronal subtypes known to control feeding and energy expenditure.1https://www.cell.com/cell-reports/fulltext/S2211-1247(19)30239-6
In this dataset, we observe GPR17 (Transcript – Blue), a G-protein-coupled receptor known to modulate glycolysis and lactate production in OLPCs,1https://www.cell.com/cell-reports/fulltext/S2211-1247(19)30239-6 at high levels in an OLPC adjacent to NPY+ INHINs. We can see genes common to many signaling pathways, including pathways downstream of lactate detection, AKT and STAT3 (Transcript – Cyan) in both GPR17+ OLPCs and adjacent NPY+ INHIN cell types.1https://www.cell.com/cell-reports/fulltext/S2211-1247(19)30239-6 Also detected are known signaling pathways downstream of GPR17 (Transcript – Yellow) in OLPCs. And with single-cell, spatial resolution researchers can study the interaction between these OLPC and INHIN cells.

Cell type map of mouse brain. Initial image depicts morphology stain of half mouse brain, which then zooms to show spatially resolved transcript location of mRNA targets overlaid with morphology stain. This fades to a spatially resolved transcript location of mRNA targets overlaid with Cell Type results, which is then enlarged to a region of interest with Cell Type results overlaid with transcripts.
Request a Quote
Contact our helpful experts and we’ll be in touch soon.
Technology Publication:
High-Plex Multiomic Analysis in FFPE Tissue at Single-Cellular and Subcellular Resolution by Spatial Molecular Imaging
References
- 1