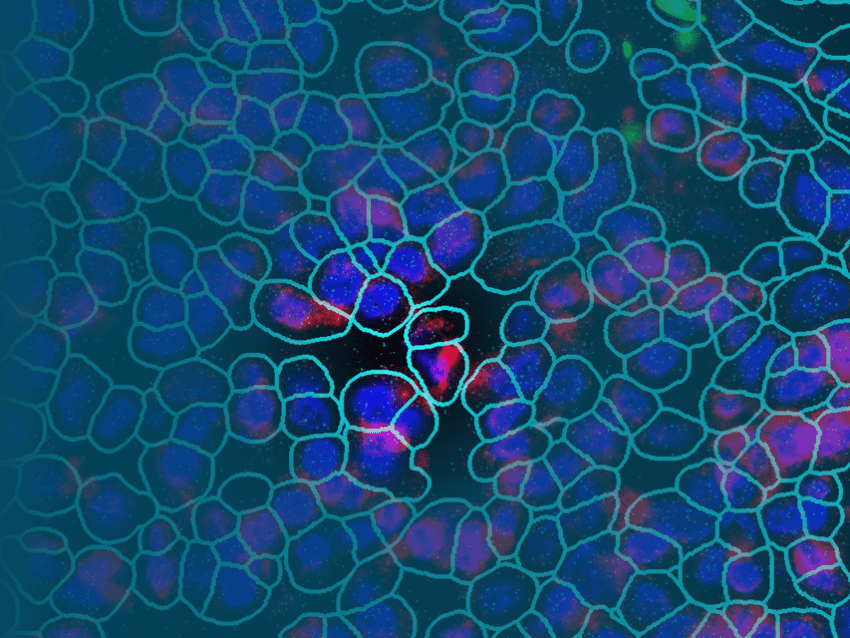
How CosMx SMI Enables Next-Generation Spatially Resolved Cell Assays with Subcellular Resolution
The ability to simultaneously map morphology, RNA, and protein at the subcellular level is the new frontier in cell biology. While the CosMx® Spatial Molecular Imager (SMI) is renowned for tissue analysis, it is also a powerhouse for in vitro applications, detecting diverse biomolecules in situ at nanometer precision. Unlike traditional dissociation methods like bulk or single cell RNA-seq that strip cells of their context, CosMx SMI enables direct subcellular spatial measurements that preserve the vital link between native morphology and microenvironmental context.
Researchers face a critical gap in high-plex, multimodal, spatially resolved assays for in vitro or ex vivo cell-based systems – particularly for drug discovery, high-throughput screening, and translational research. Legacy tools force a compromise: you get either molecular depth or spatial fidelity, but rarely both. CosMx SMI eliminates this trade-off by delivering whole-transcriptome coverage, and protein co-detection in a scalable, cost-effective framework.
This introduces a new paradigm: spatially resolved cell assays that integrate morphology, multiomics, and subcellular biology in situ.
- CosMx Assays with On-Slide Cultured Cells
- CosMx Assays with Deposited Cells
- CosMx Assays with Auto-Dispensed (Nano-Printed) Cell Arrays
- The Future of Cell Assays in Spatial
CosMx Assays with On-Slide Cultured Cells
Culturing cells directly on slides maintains the native cellular architecture essential for functional studies (Fig. 1). This configuration supports Morpho-Transcriptomics –integrating cellular morphology with high-plex molecular readouts (1).
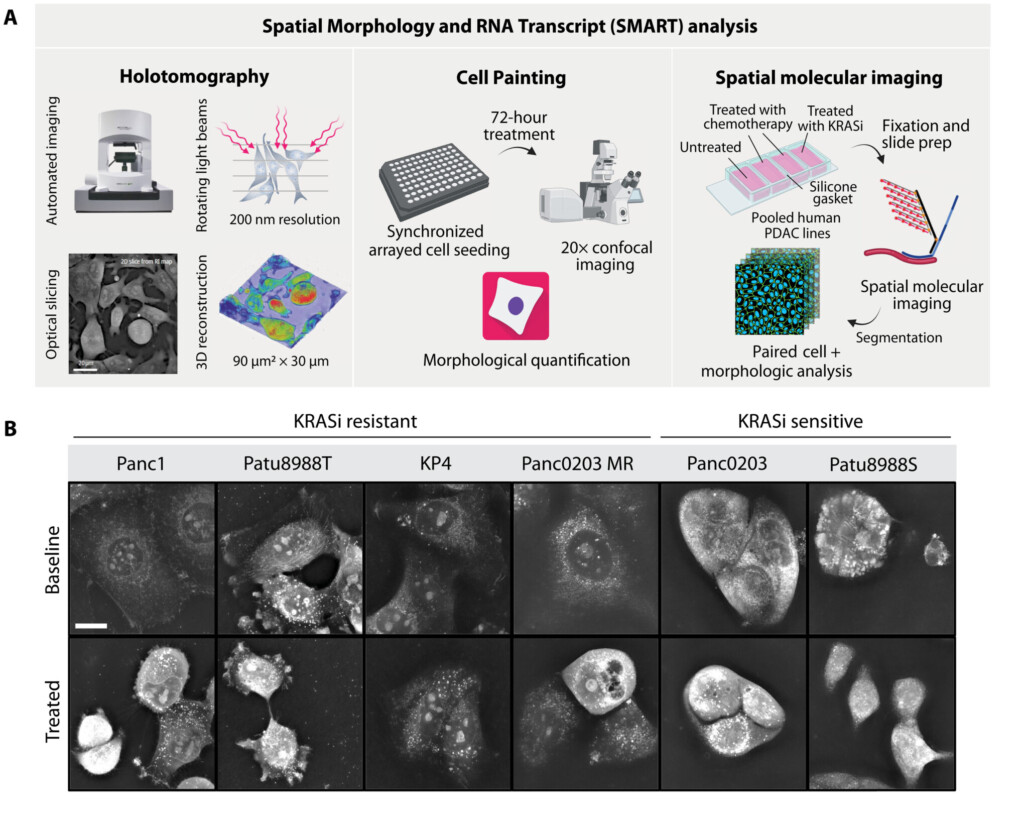
Crucially, CosMx SMI enables organelle-resolved profiling, where transcripts and proteins can be localized within the nucleus, cytoplasm, or other subcellular compartments (Fig. 2) (2). This capability enables the discovery of regulatory processes such as mRNA localization, nuclear export, RNA-protein interactions, and spatially restricted signaling cascades (Fig. 3). Importantly, the ability to maintain morphological fidelity allows direct, high confidence correlation between cell shape, cytoskeletal architecture, and molecular states – a feature typically lost in dissociation-based assays.
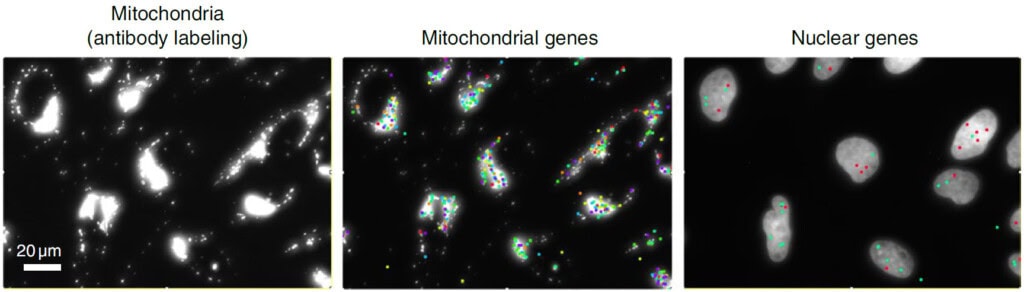
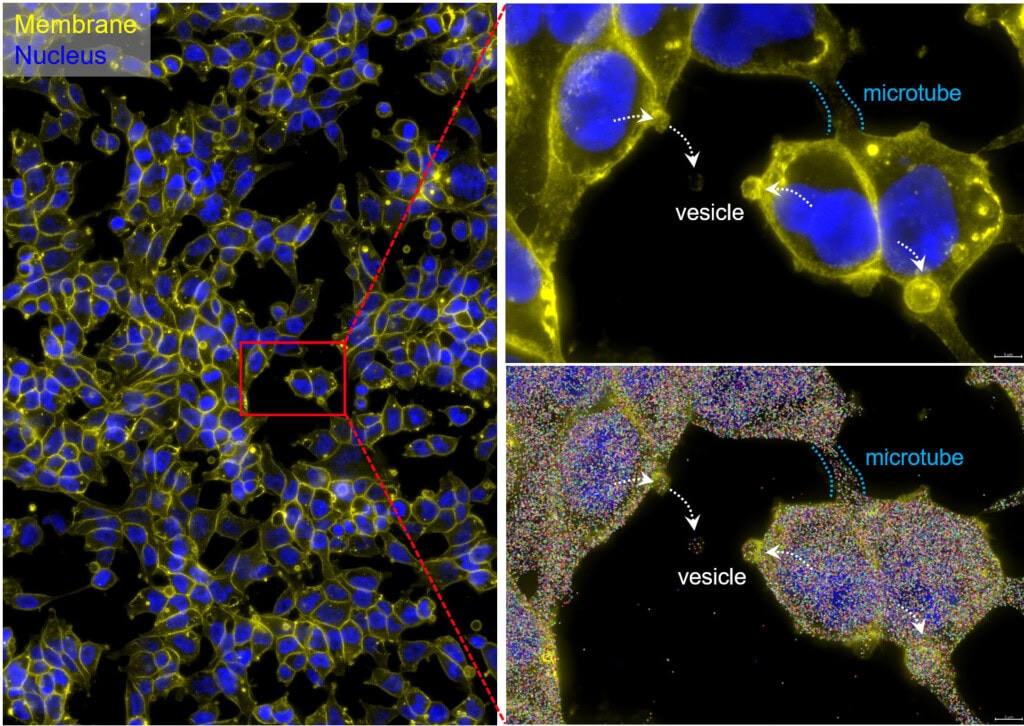
An on-slide culture workflow is uniquely valuable for adherent cell types, such as epithelial or neuronal cells, where morphology is crucial for function. By maintaining cells in their native state, this method minimizes artifacts caused by cell dissociation and enables powerful longitudinal studies of cellular dynamics. The high-plex capacity of CosMx SMI further allows simultaneous profiling of thousands of RNA transcripts and dozens of proteins, providing a comprehensive, multiomic view within morphologically intact cells (Fig. 4) (3).
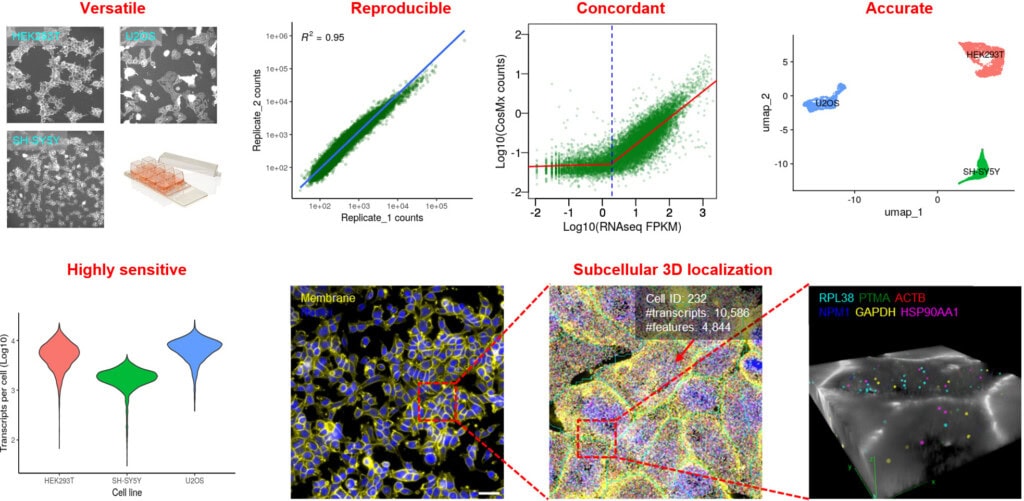
Protocol Tips – To accommodate the unique properties of cultured cells, we developed a dedicated protocol with key changes to the standard CosMx sample preparation workflow:
- Use Poly-L-Lysine precoated slides (e.g., Electron Microscopy Sciences Cat# 63410-02) for optimal cell adherence
- The recommended initial seeding concentration is 100,000 cells/mL to ensure mono-dispersion of cells on slide
- A variety of on-slide removable chambers can be used (e.g., labware from ibidi, Grace Bio-Labs, LabTek), depending on experimental design and throughput requirement
- Once cells reach 70% confluence, fix them with fresh 10% NBF or 4% PFA for 30 min at room temperature
- Skip the target retrieval step in the sample preparation
- Use a milder, selective digestion enzyme (e.g., 5 µg/mL endoproteinase GluC) instead of proteinase K
- Reduce the use of fiducials by 1/2 – 2/3 (e.g., 0.0005% fiducials in cell-based assays vs. 0.001% in standard CosMx sample preparation)
Detailed protocol for CosMx cultured cell assays can be found at:
- Sub-cellular Imaging of the Entire Protein-Coding Human Transcriptome (18933-plex) on FFPE Tissue Using Spatial Molecular Imaging
- Integrated spatial morpho-transcriptomics predicts functional traits in pancreatic cancer
CosMx Assays with Deposited Cells
For non-adherent or suspension cells, such as PBMCs, CosMx assays can be adapted to analyze cells deposited onto slides with subcellular spatial information preserved. A notable example is the Spatial Transcriptomics Analysis by Multiplexed Profiling (STAMP) method (Pitino et al.), which integrates CosMx SMI with cell deposition techniques to enable high-throughput spatial profiling of single cells.
In this workflow, cells are deposited onto poly-L-lysine coated slides (for multiple samples, assemble a multi-well chamber on CosMx slide using Grace Bio-Labs ProPlate chambers, ibidi µ-Slide or isolator gaskets), preserving spatial integrity for single-cell analysis. This allows researchers to map the spatial distribution of RNA and proteins within these cells (e.g., utilizing the ratio of nuclear and cytoplasmic transcripts for RNA velocity modeling, check Analyzing genes’ subcellular localization – Blog), facilitating studies on cellular heterogeneity and biomolecular localization in complex populations.
This strategy provides a platform for population-scale analyses across a considerable number of conditions, cell lines, or perturbations. By combining CosMx SMI with deposition-based workflows, researchers can achieve spatially-resolved, parallelized, scalable, and reproducible assays – ideal for screening, intracellular phenotypic characterization, and drug mechanism studies.
Detailed STAMP protocol can be found at:
CosMx Assays with Auto-Dispensed (Nano-Printed) Cell Arrays
For ultra-high-throughput screening, CosMx cell-based assays can be scaled using nano-liter dispensing systems to construct high-density cell arrays on slides. This workflow uses precise dispensing of cell suspensions onto CosMx-compatible slides to create arrays of single cells or small cell clusters without the need for chambers or isolators (Fig. 5). Each array spot acts as an independent assay unit, ideal for screening applications, such as drug response studies, where large numbers of conditions must be analyzed rapidly and cost-effectively.
The auto-dispensing workflow capitalizes on the multiomic capabilities of CosMx SMI to profile molecular responses across diverse cell types or treatment conditions. For example, researchers can use this approach to screen cancer cell lines for drug sensitivity, mapping transcriptomic and proteomic changes at subcellular resolution across hundreds of conditions in a single experiment. The spatial resolution of CosMx SMI further allows for the analysis of intracellular responses, such as changes in organelle-specific gene expression, providing a granular view of drug mechanisms. By combining high-throughput array construction with high-plex profiling, this workflow represents a powerful tool for precision medicine and systems biology.
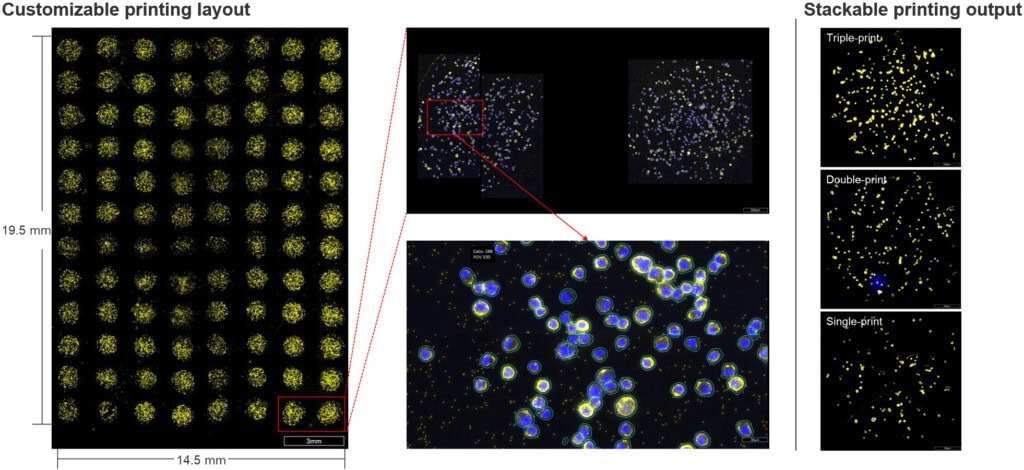
Protocol Tips: To accommodate the unique properties of nano-printed cells, we recommend the following:
- Fix the harvested cell samples with fresh 10% NBF or 4% PFA. For long-term storage, cells can be put in 10% DMSO-containing buffer or culture medium at -80°C.
- Begin with a cell concentration of 106 cells/mL for dispensing, corresponding to 1 cell/nL
- Recommended dispensing volume of 100 nL, giving rise to a spot size of 700-800 µm
- The dispensed droplets can be quickly dried with a temperature gradient (25°C, 10min → 42°C, 10min → 22°C, 10min), followed by incubation with 70% ethanol at room temperature (2 hours) or 4 °C (up to 1 month)
- The sample preparation for RNA assays follows the established cultured cell workflow, where 0.2 µg/mL ProK is used together with 5 µg/mL GluC to achieve optimal digestion

Analyzing High-throughput Subcellular Data from CosMx Assays
To unlock the full potential of high-throughput subcellular data, below are few computational tools that could be used:
- CellSP: A computational framework that identifies, visualizes, and characterizes recurring subcellular spatial “gene-cell modules” – gene sets with coordinated transcript localization across cells (5).
- BENTO: A Python toolkit within the Scverse ecosystem that ingests single-molecule coordinate data and segmentation masks to define subcellular domains, annotate localization patterns, and quantify gene-gene co-localization (6).
- ELLA: A robust, scalable tool designed to model and quantify subcellular spatial variation of gene expression at high resolution across individual cells (7).
- InSTAnT: A toolkit for extracting molecular relationships and subcellular localization patterns of RNA, such as cellular domain-specific gene pairs, from single-molecule resolution data like CosMx SMI (8).
The Future of Cell Assays in Spatial
CosMx SMI transforms cell-based assays from simple counts to complex spatial narratives. By preserving spatial context, enabling high-plex multimodal profiling, and supporting high-throughput formats, these assays unlock new possibilities for studying cellular and subcellular biology. By integrating morpho-transcriptomics, organelle-specific omics, and scalable screening, we are opening new avenues for precision medicine and drug mechanism studies.
Looking ahead, the future involves coupling CosMx SMI with machine learning to streamline complex spatial data interpretation and integrating it with live-cell imaging or CRISPR screens for dynamic, longitudinal insights into cellular responses. CosMx spatially resolved cell-based assays open new avenues for drug discovery, biomarker development, and mechanistic studies by linking perturbations to spatial multiomic signatures at single-cell resolution. They enable organelle-level profiling, integration of morphology with molecular states, and AI-driven phenotype discovery, offering a powerful alternative to traditional high-throughput assays.
References
- Gong, D. et al. “Integrated Spatial Morpho-Transcriptomics Predicts Functional Traits in Pancreatic Cancer.” Science Advances 2025, Vol 11, Issue 42 doi: 10.1126/sciadv.adx0632.
- He, S. et al. “High-Plex Imaging of RNA and Proteins at Subcellular Resolution in Fixed Tissue by Spatial Molecular Imaging.” Nat Biotechnol, vol. 40, no. 12, 2022, pp. 1794-806, doi:10.1038/s41587-022-01483-z.
- Khafizov, R. et al. “Sub-Cellular Imaging of the Entire Protein-Coding Human Transcriptome (18933-Plex) on Ffpe Tissue Using Spatial Molecular Imaging.” bioRxiv, 2024, doi:https://doi.org/10.1101/2024.11.27.625536.
- Pitino, E. et al. “Stamp: Single-Cell Transcriptomics Analysis and Multimodal Profiling through Imaging.” Cell, vol. 188, no. 18, 2025, pp. 5100-17.e26, doi:10.1016/j.cell.2025.05.027.
- Aggarwal, B. and S. Sinha. “Cellsp: Module Discovery and Visualization for Subcellular Spatial Transcriptomics Data.” bioRxiv, 2025, doi:10.1101/2025.01.12.632553.
- Mah, C. K. et al. “Bento: A Toolkit for Subcellular Analysis of Spatial Transcriptomics Data.” Genome Biol, vol. 25, no. 1, 2024, p. 82, doi:10.1186/s13059-024-03217-7.
- Wang, J. X. and X. Zhou. “Ella: Modeling Subcellular Spatial Variation of Gene Expression within Cells in High-Resolution Spatial Transcriptomics.” bioRxiv, 2024, doi:10.1101/2024.09.23.614515.
- Kumar, A. et al. “Intracellular Spatial Transcriptomic Analysis Toolkit (Instant).” Nat Commun, vol. 15, no. 1, 2024, p. 7794, doi:10.1038/s41467-024-49457-w.