GeoMx DSP: What’s in an ROI?
In the field of medical imaging, the region of interest (commonly abbreviated as ROI) is an area within an image that is selected for a particular purpose, including defining boundaries or measuring the size of a feature such as a tumor. Similarly, in terms of imaging for spatial biology, the ROI is the area selected within a tissue sample to measure gene or protein expression.
Spatial multiomics allows the study gene or protein expression at high resolution within intact tissue, while maintaining positional information in a particular tissue region or within a single cell population. Accurate placement of ROIs greatly influences the quality of data and the ability to answer the given research question. For instance, tumors are not uniform masses of cells, but consist of subpopulations of cells, with distinct genotypes and phenotypes that may harbor divergent biological behaviors. Hence, the analysis of a subpopulation of cells within a tumor heavily depends on the choices taken during ROI selection.
ROI Selection with GeoMx DSP
 The GeoMx DSP Instrument
The GeoMx DSP InstrumentThere are several spatial multiomics methods available, and each differs in the way they capture and store spatial information. NanoString’s GeoMx® Digital Spatial Profiler (DSP) is a histology-driven spatial multiomics platform that profiles mRNA as well as proteins within a user-defined ROI based on the appropriate morphology markers for the tissue. Subsequently, profiling data is collected from the ROI instead of the whole tissue.
To measure gene and protein expression, the GeoMx DSP utilizes a digital barcoding technology where oligo-barcodes are attached via a photocleavable linker to fluorescent antibodies or RNA in situ hybridization probes. A pool of these barcode-labeled probes is then hybridized to mRNA or protein targets on a tissue section. This is followed by either selecting single or multiple ROIs in the desired location on the tissue section. These ROIs are then exposed to UV light that causes the release of the DSP barcodes which are then collected and sequenced through standard NGS procedures or counted using the nCounter® Analysis System. The identity and quantity of the collected barcodes can then be translated into expression amounts of transcripts and proteins and mapped back digitally to the ROI within the tissue section.

Flexible and Customizable ROI Selection
One of the major advantages of the GeoMx DSP platform is the flexibility with which you can select regions of the tissue to profile; other transcriptomics platforms (such as Visium) capture data non-specifically from the entire tissue section in an array-shaped pattern.
This flexible ROI strategy is possible due to the programmable digital micromirror device (DMD) which is a key feature of the GeoMx DSP technology.1Beechem J.M. High-Plex Spatially Resolved RNA and Protein Detection Using Digital Spatial Profiling: A Technology Designed for Immuno-Oncology Biomarker Discovery and Translational Research. Methods Mol. Biol. 2020;2055:563–583 DMDs are small electromechanical systems that contain an array of steerable reflective micromirrors (~2 million total mirrors) that can be dynamically configured to profile any shape ROI on the tissue and maximize the biological information obtained from it. Therefore, the ROI can be of any shape or size from within the field of view, including non-contiguous regions, such as a particular cell type population.
With GeoMx DSP, the ROI can be of any shape or size from within the field of view — including non-contiguous regions.
The ROI shapes can be either regular geometric shapes or irregular shapes with sizes ranging from 10 to 600 μm in diameter. Although the ROIs can be as small as 10 μm — the size of a single cell — the GeoMx throughput is not conducive to single-cell analysis: you need at least 20 to 300 cells per ROI for reliable quantification of transcripts and proteins.
These ROIs can be selected in different areas of the tissue defined by certain spatial locations and/or morphological markers. The UV light is shone over a mask pattern formed by the DMD and can be focused on the area stained by a particular morphology marker within an ROI, thus further subdividing that ROI into a segmented area of illumination (AOI) that can be profiled separately. Various sizes and types of AOIs are also possible, from a minimum region size of 5 µm × 5 µm to a maximum of 660 µm × 785 µm. UV light directed via the DMD illuminates all selected ROIs sequentially. As a result, the indexing oligos are released only within the boundaries of the selected ROI. Consequently, the spatial resolution of the GeoMx DSP depends on the ROI selection strategy and sampling plan.
Strategies to define an ROI
The GeoMx DSP allows for various ROI selection strategies, including placing geometric shapes around specific areas (“geometric”), drawing concentric circles surrounding a histological landmark (“contour”), gridding across the entire tissue section (“gridded”), and segmentation within an ROI (“segmented”).2Beechem J.M. High-Plex Spatially Resolved RNA and Protein Detection Using Digital Spatial Profiling: A Technology Designed for Immuno-Oncology Biomarker Discovery and Translational Research. Methods Mol. Biol. 2020;2055:563–583.
Bergholtz, H., Carter, J. M., Cesano, A., U Cheang, M. C., Church, S. E., Divakar, P., Fuhrman, C. A., Goel, S., Gong, J., Guerriero, J. L., Hoang, M. L., Hwang, E. S., Kuasne, H., Lee, J., Liang, Y., Mittendorf, E. A., Perez, J., Prat, A., Pusztai, L.,… Cancer Consortium, G. B. (2021). Best Practices for Spatial Profiling for Breast Cancer Research with the GeoMx® Digital Spatial Profiler. Cancers, 13(17). https://doi.org/10.3390/cancers13174456
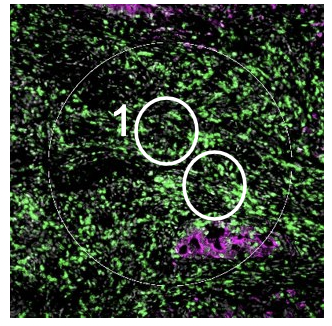
Geometric Profiling
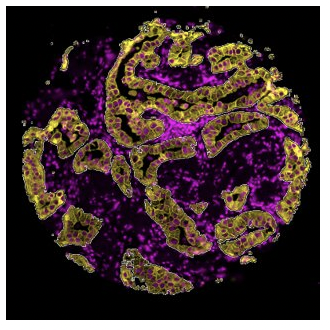
Segmentation

Cell Type Specific
What is the expression profile of a specific cell population in my tissue?
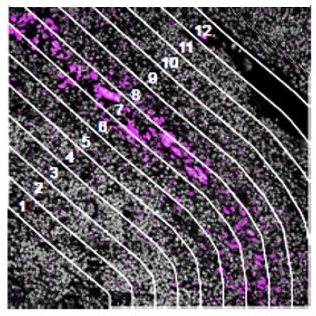
Contour
How does the immune environment change on either side of an infiltrate boundary?
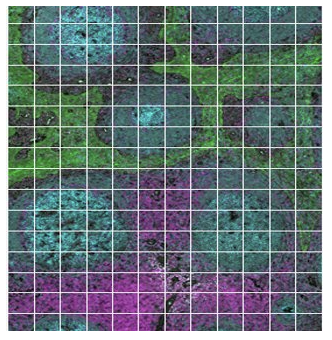
Gridded
What novel targets are uncovered with deep mapping of a specific tissue region?
The geometric profiling strategy offers standardized geometric shapes or custom polygons and is particularly useful to profile large tissue areas or measure tissue heterogeneity at different locations for example within and near to a tumor. The gridded profiling strategy expands on the geometric profiling concept by placing geometric ROIs at regularly defined intervals across the tissue sample, especially useful for an unbiased analysis. Contour profiling is particularly useful to measure gradients and distances around a biological feature to examine how proximity affects a biological response. In this case, the biological feature of interest is placed within an ROI, then AOIs at fixed distances from that feature are serially illuminated and the released oligos individually collected.
The segmented profiling strategy is another simple method that allows users to define an ROI which can be then further subdivided or segmented into more Areas of Interest (AOIs). For example, segmented profiling has been used to assess protein expression in separate tissue compartments in a tumor biopsy: the tumor cells and the surrounding microenvironment consisting of stroma and immune cells.3Song, X., Xiong, A., Wu, F., Li, X., Wang, J., Jiang, T., Chen, P., Zhang, X., Zhao, Z., Liu, H., Cheng, L., Zhao, C., Wang, Z., Pan, C., Cui, X., Xu, T., Luo, H., & Zhou, C. (2022). Original research: Spatial multi-omics revealed the impact of tumor ecosystem heterogeneity on immunotherapy efficacy in patients with advanced non-small cell lung cancer treated with bispecific antibody. Journal for Immunotherapy of Cancer, 11(2). https://doi.org/10.1136/jitc-2022-006234 A tumor cell marker, Pancytokeratin (PanCK), was used to guide the selection of AOIs within a given ROI. PanCK-expressing regions within the ROI were exposed to UV light and collected for analysis, followed by UV illumination of the tumor inverse area, or PanCK-negative area, within the same ROI. A heatmap of tumor (PanCK-positive) and tumor inverse segments (PanCK-negative) revealed that two distinct expression profiles, with a strong enrichment of immune markers in the tumor inverse or stromal compartment, providing insights into crosstalk between tumor cells and immune cells in response to therapy.
A variation of the segmentation strategy is cell type-specific profiling that characterizes expression changes within a specific cell type population that are not necessarily contiguous.
One GeoMx experiment may involve a series of several or all the aforementioned ROI selection strategies, depending on the questions being asked.
There is no wrong way or right way to develop your ROI selection strategy in a GeoMx experiment; it depends on the types of questions you are looking to answer. One GeoMx experiment may involve a series of several or all the aforementioned ROI selection strategies, depending on the questions being asked and/or the hypothesis. This flexibility truly makes the GeoMx DSP a unique instrument for spatial biology.
Best of all, the GeoMx DSP technology is non-destructive, meaning that you can save the sample after the experiment — unlike laser capture microdissection, which removes a given region from the tissue and destroys the sample in the process.
References
- 1Beechem J.M. High-Plex Spatially Resolved RNA and Protein Detection Using Digital Spatial Profiling: A Technology Designed for Immuno-Oncology Biomarker Discovery and Translational Research. Methods Mol. Biol. 2020;2055:563–583
- 2Beechem J.M. High-Plex Spatially Resolved RNA and Protein Detection Using Digital Spatial Profiling: A Technology Designed for Immuno-Oncology Biomarker Discovery and Translational Research. Methods Mol. Biol. 2020;2055:563–583.
Bergholtz, H., Carter, J. M., Cesano, A., U Cheang, M. C., Church, S. E., Divakar, P., Fuhrman, C. A., Goel, S., Gong, J., Guerriero, J. L., Hoang, M. L., Hwang, E. S., Kuasne, H., Lee, J., Liang, Y., Mittendorf, E. A., Perez, J., Prat, A., Pusztai, L.,… Cancer Consortium, G. B. (2021). Best Practices for Spatial Profiling for Breast Cancer Research with the GeoMx® Digital Spatial Profiler. Cancers, 13(17). https://doi.org/10.3390/cancers13174456 - 3Song, X., Xiong, A., Wu, F., Li, X., Wang, J., Jiang, T., Chen, P., Zhang, X., Zhao, Z., Liu, H., Cheng, L., Zhao, C., Wang, Z., Pan, C., Cui, X., Xu, T., Luo, H., & Zhou, C. (2022). Original research: Spatial multi-omics revealed the impact of tumor ecosystem heterogeneity on immunotherapy efficacy in patients with advanced non-small cell lung cancer treated with bispecific antibody. Journal for Immunotherapy of Cancer, 11(2). https://doi.org/10.1136/jitc-2022-006234