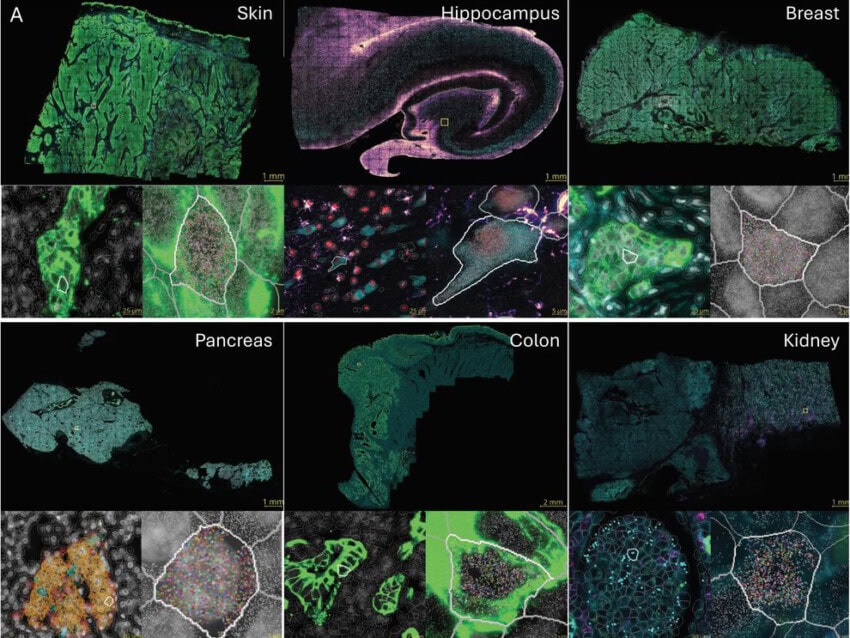
Moving Beyond scRNA-seq: The Power of CosMx Whole Transcriptome
Single-cell RNA sequencing (scRNA-seq) has transformed molecular biology by enabling high-throughput transcriptional profiling at cellular resolution. But as powerful as scRNA-seq is, it comes with trade-offs: loss of spatial information, sample dissociation artifacts, and incomplete cell representation, especially for rare or irregularly shaped cell types.
The CosMx® Spatial Molecular Imager (SMI) offers a new paradigm for single-cell biology: combining whole-transcriptome coverage with subcellular spatial resolution using intact FFPE or fresh frozen tissue. The new CosMx Whole Transcriptome (WTX) panel targets 18,942 human genes using cyclic hybridization and high-resolution imaging, enabling full transcriptome profiling with subcellular resolution and without sequencing.
Here, we compare CosMx WTX vs scRNA-seq in head-to-head analyses and highlight where spatial transcriptomics is redefining what’s possible in single-cell biology.
Information Losses from Tissue Disruption
The dissociation step required for scRNA-seq is inherently and necessarily disruptive. Cells must be enzymatically or mechanically separated, introducing technical variability and often resulting in the loss of fragile, rare, or structurally complex cell types. CosMx bypasses this entirely by imaging intact tissue, preserving both morphology and molecular content. The architectural context is scientifically essential for understanding how cells interact and organize within tissue microenvironments.
In a poster presented at AGBT 2025, we completed a direct comparison of droplet-based scRNA-seq and CosMx WTX using adjacent FFPE sections from colorectal carcinoma and pancreas samples. The CosMx WTX panel profiled over 400,000 cells per section—up to 100 times more cells than scRNA-seq from the same tissue blocks (which typically captured fewer than 10,000 cells). This level of throughput not only enhances statistical power but also enables robust characterization of spatially organized cellular niches.
CosMx SMI also preserved epithelial and muscle cells that were underrepresented in scRNA-seq due to their dense packing or irregular shape. For studies where every cell matters—such as profiling the tumor microenvironment or mapping stem cell niches—this level of fidelity is essential.
Summary table of head-to-head comparison from FFPE colorectal samples.
(From Cui et al., 2025.)
| Performance metrics | scRNA-seq | CosMx WTX |
| Total number of genes | 18,082 | 18,935 |
| Input sample area | N/A | 103.63 mm2 |
| Input number of cells | 10,000 | 493,929 |
| Output number of cells | 6,275 | 493,929 |
| Median transcripts per cell | 1,151 | 967 |
| Median unique genes per cell | 774 | 627 |
| Genes above background | N.A. | 13,918 |
| Genes with high dynamic range | 3,349 | 5,896 |
| Coverage of tissue composition | Biased | Unbiased |
Amplification Bias: A Hidden Variable in scRNA-seq
Traditional scRNA-seq workflows rely on reverse transcription and PCR amplification to convert RNA into cDNA, a process that can introduce significant bias. Amplification efficiency varies across transcripts due to factors like GC content and secondary structure, leading to uneven representation of gene expression levels. While unique molecular identifiers (UMIs) can be employed to mitigate this issue, they cannot fully correct for the distortions introduced during amplification.
In contrast, the CosMx SMI utilizes a direct hybridization approach without the need for enzymatic amplification, thereby preserving the original transcript abundance. This amplification-free methodology ensures a more accurate and reproducible quantification of gene expression, particularly beneficial when analyzing low-abundance transcripts or working with degraded samples like FFPE tissues.
Rare Cell Types Finally Captured
One of the persistent blind spots of scRNA-seq is the recovery of low-abundance or morphologically distinct cells. In a human pancreas tissue, CosMx WTX detected ~130 endocrine Epsilon cells, which are responsible for ghrelin production, from out of nearly 400,000 total cells. These cells constitute less than 0.05% of the pancreas population and were entirely missed by scRNA-seq in the same experiment.
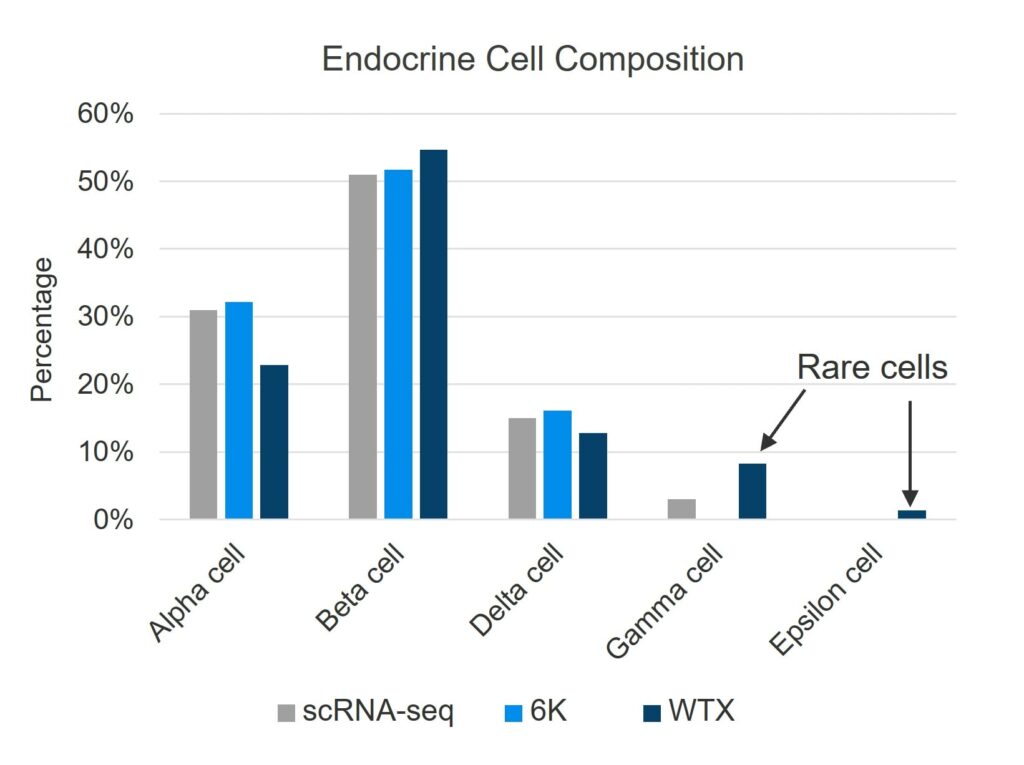
Cell types detected from human pancreas, analyzed via three different methods: droplet-based scRNA-seq, CosMx Human 6K Discovery Panel, and CosMx Human WTX Panel. Only the CosMx WTX workflow detected the rarest Epsilon cell type. (From Cui et al., 2025.)
Comparable Gene Detection with More Spatial Precision
With respect to transcript detection, the CosMx WTX panel was comparable to scRNA-seq. As shown in the earlier Summary Table, both the CosMx platform and droplet-based scRNA-seq detected 18,000-19,000 genes from FFPE colon tissue. Median transcript counts per cell and unique gene counts were also similar across both platforms.
However, the CosMx platform brings valuable information that scRNA-seq cannot: spatial localization. Each transcript is assigned to a precise subcellular position, allowing for in situ identification of rare cell types, states, and interactions. With the high coverage of WTX, researchers can now map ligand-receptor relationships or pathway activity directly onto the tissue architecture. Learn more about pathways-first biology here.
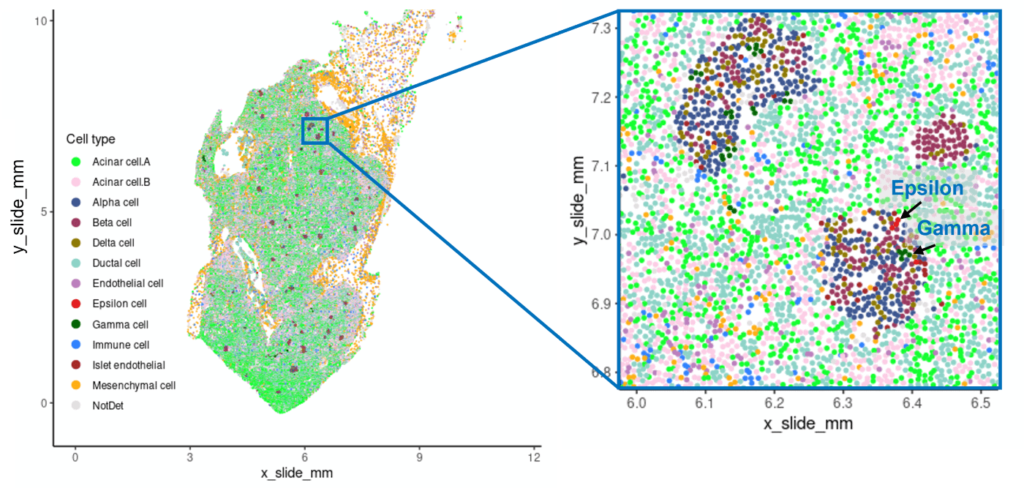
Localization of human pancreas cell types detected via CosMx WTX, with rare Gamma and Epsilon types indicated by arrows. (From Cui et al., 2025.)
Advanced Analysis Tools Bring the Whole Biological Context
Supporting the output of CosMx SMI is the AtoMx® Spatial Informatics Platform, which enables powerful, code-free analysis and visualization of spatial transcriptomic data. The AtoMx platform features an AI-driven multi-modal segmentation algorithm that accurately defines single cells using multiplex protein and nuclear markers. With tools like Insitutype, users can classify cell types and visualize their distribution across the tissue landscape. The platform also supports advanced spatial analyses including neighborhood mapping to define tissue niches and cell–cell interfaces. Researchers can interrogate expression of individual transcripts at subcellular resolution, a level of precision not achievable with grid- or array-based methods.
A New Paradigm in Tissue Research
While scRNA-seq remains a valuable tool for discovery in dissociated cell systems, many biological research questions demand more. CosMx WTX offers a more complete approach, bringing spatial resolution, cell-cell interaction mapping, and comprehensive cell recovery from FFPE tissue.

Summary of the advantages of CosMx SMI over scRNA-seq.
With CosMx WTX, researchers can move beyond transcript lists and clustering maps to directly observe how the entire transcriptome is expressed in space. This technology “holy grail” enables advanced biological approaches from cell-cell interactions to pathway mapping—analyses with which dissociation-based approaches simply cannot compete.
Further Reading
Insitutype: likelihood-based cell typing for single cell spatial transcriptomics

Related Content