Tips to optimize the CosMx Multiomics sample prep for fragile tissues
Tips to optimize the CosMx Multiomics sample prep for fragile tissues
The CosMx multiomics platform enables sequential protein and RNA analysis on the same tissue sample, combining a 64-plex protein panel with a high-plex RNA panel (e.g., the whole-transcriptome RNA panel). This document outlines best practices for fragile samples during this sequential workflow. .
Sequential Readout Workflow Overview

Figure. 1 Schematic of the CosMx same-sample multiomics workflow
- Protein Assay: A 64-plex protein panel is applied for IO protein mapping. Unique to the multiomics workflow, antibodies are temporarily fixed in situ using a reversible fixative Dithiobis(succinimidyl propionate) (DSP), which is removed post-data collection with a reducing reagent Tris-(2-Carboxyethyl)phosphine (TCEP). The 64-plex protein panel is also run using a newly introduced removable flow cell coverslip in place of the standard flow cell coverslip.
- RNA Assay: After the protein run, the CosMx slide is retrieved from the instrument, and the flow cell coverslip is removed. A high-plex RNA panel is then hybridized to the same sample for transcriptomic analysis and a new flow cell coverslip is applied for the RNA part of the instrument run.
The Prevalence of Multiomics Workflow Related Cell Loss
In the CosMx multiomics workflow, the tissue sample may experience additional mechanical stress due to increased handling and liquid exchange. For especially fragile tissue samples, cell and/or tissue loss can occur before or during the RNA run, impacting data quality. In general, it’s common to observe about 5-10% cell loss between the protein run and the RNA run. Notably, we observe certain tissue features (e.g. adipose, connective stroma) being more susceptible to the multiomics workflow related cell loss.
- The following tissue types have been evaluated and proven compatible with the multiomics workflow: Brain, Breast, Colon, Kidney, Liver, Lung, Pancreas, Skin, Tonsil, Thymus, Fallopian tubes.
- Tissues with lower cellular content or loose structures, such as adipose, airways, or connective stroma, are more fragile in general. In general, non-cancerous tissues (e.g., skin, colon) may be more vulnerable to multiomics workflow related cell loss.
- Tissues with highly packed cells or delicate structures where mono-layer sections are challenging to obtain, such as thymus, may experience more cell loss in dense regions.
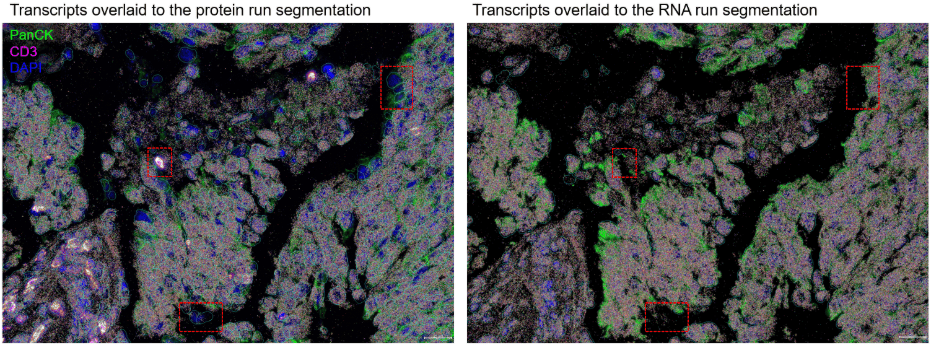
Figure 2. Multiomics workflow related cell loss in a WTX multiomics run. Cells located in stromal regions or at the edge of tissue regions (highlighted with red boxes) may be more prone to loss.
How to Assess Multiomics Workflow Related Cell Loss
Tissue types or blocks previously validated with a CosMx RNA panel are generally well-suited for CosMx multiomics assays. The acceptable threshold for cell loss should be defined based on experimental design or project requirements. Effective evaluation of cell loss is critical for ensuring data quality. The multiomics workflow related cell loss can be effectively evaluated in 2 major ways.
- Assess Tissue Integrity: Use upfront morphology images to evaluate tissue loss, focusing on immune-enriched regions (e.g., CD45+ or CD68+ cell populations).
- Compare Cell Counts: Quantify cell numbers from protein and RNA segmentation outputs prior to dataset integration to identify discrepancies indicative of cell loss.
Optimize Sample Preparation for Fragile Tissues
To ensure high-quality data in CosMx multiomics assays, careful sample preparation is essential, particularly for fragile tissues prone to cell loss during sequential protein and RNA readouts. The following guidelines outline optional digestion conditions that can be used to balance tissue integrity and assay performance, with adjustments based on observed cell loss.
- Standard Digestion: Use 3 µg/mL proteinase K in 1× PBS for sample preparation.
- Modified Digestion for High Cell Loss: If tissue or cell loss exceeds 15% before the RNA run, switch to a milder digestion treatment:
- Optimized Digestion: 1 µg/mL proteinase K in 0.5% PBST (Tween-20 in 1× PBS).
- Rationale: A lower Proteinase K concentration minimizes tissue disruption while ensuring sufficient permeabilization for RNA hybridization and readout. The addition of a non-ionic detergent (Tween-20) promotes homogeneous digestion, reducing local steric hindrance for reagent delivery.
General Recommendations
Careful attention to quality control, sample handling, and documentation is essential to maximize the success of a CosMx multiomics assay. These best practices guidelines can help to mitigate cell loss and ensure consistent data quality across sequential protein and RNA readouts, particularly for fragile tissue samples.
- Quality Control: After the protein run and flow cell removal, inspect the sample for integrity. If tissue or cell loss exceeds 15%, pause and adjust the protocol as described.
- Sample Handling: Minimize mechanical stress during slide retrieval and flow cell removal to preserve tissue architecture.
- Documentation: Record cell loss percentages and protocol modifications for each sample to refine workflows for specific tissue types.
Conclusion
By implementing these best practices, researchers can reduce cell loss in fragile tissue samples during the CosMx multiomics assay, ensuring high-quality protein and RNA data. For fragile tissues like non-cancerous skin and colon, early adoption of milder digestion protocols is recommended. Always monitor cell loss thresholds and adjust workflows as needed to optimize results.