Breakthroughs in Spatial Biology: AGBT 2025 Highlights
AGBT 2025 was a whirlwind of innovation and excitement. Our Morning Buzz Sessions provided deep dives into the latest in spatial biology each day, while our Brews with Bruker gatherings served up lively conversation over craft brews. We also hosted a Silver Sponsor Workshop, where experts led in-depth discussions across all Bruker platforms. Whether you joined us in person or are catching up from afar, check out our e-book with a collection of posters and a link to view our workshop presentation and stay caught up on our latest scientific breakthroughs.
Streamlining Imaging-Based Spatial Biology with EpicIF– A Universal Signal Removal Strategy
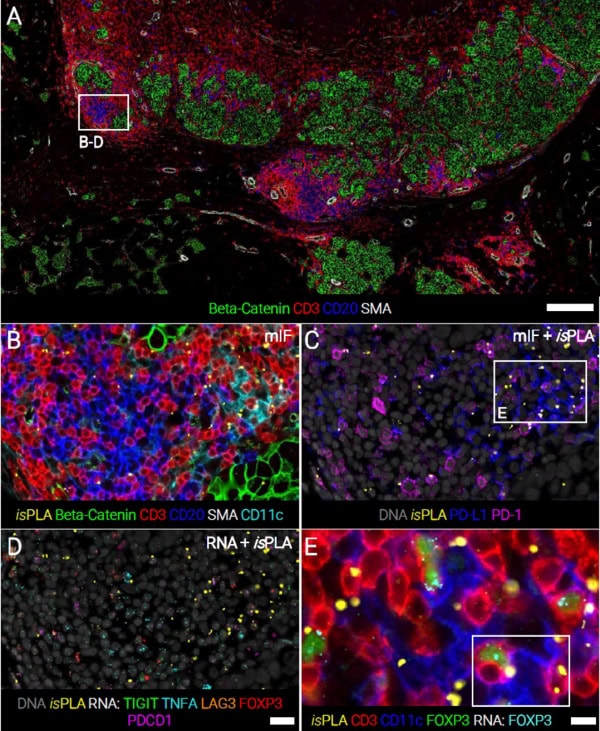
Check out how EpicIF™ technology for CellScape™ Precise Spatial Proteomics enables flexible, high-plex immunofluorescence on the same tissue section, removing existing fluorescence signals after each imaging cycle to streamline multi-omic assays. This poster couples iterative protein immunofluorescence, RNA-FISH, and an in situ proximity ligation assay (isPLA) on a single FFPE slide, pinpointing not only protein and transcript levels but also direct protein–protein interactions (PD-1/PD-L1). We illustrate this with an invasive ductal carcinoma sample, profiling tumor and immune cells (including tertiary lymphoid structures) to reveal nuanced immunoregulatory processes in the tumor microenvironment.
CosMx SMI Spatially Resolved Whole Transcriptome in FFPE Tissue – A Paradigm Shift for Tissue Analysis
See how our brand new CosMx® Whole Transcriptome (WTX) technology overcomes the limitations of tissue dissociation and single-cell capture in conventional single-cell RNA sequencing (scRNA-seq). This poster showcases the profiling of adjacent FFPE sections in parallel with droplet-based scRNA-seq, demonstrating that CosMx WTX can detect a highly comparable set of genes while fully preserving spatial context. Additionally, we capture extremely rare cell types commonly lost in dissociation-based methods. We also highlight the workflow’s exceptional throughput, recovering hundreds of thousands of cells and delivering more comprehensive data than scRNA-seq alone.
InSituDiff: Disease-Driven Changes in Cellular Neighborhoods as a Window into Spatial Transcriptomics
Learn a new cell segmentation approach that supports data analysis for the CosMx WTX panel. This poster introduces a new cell segmentation approach tailored to the intricate shapes found in brain tissue, which is especially useful when performing high-coverage spatial research with CosMx WTX. By training a specialized neural network on diverse neuronal cell types and employing a “high-recovery” post-processing method, we show how our pipeline captures challenging star-like and elongated cells that more generic models often miss. In practical tests on FFPE human brain samples, the improved segmentation recovers more transcripts per cell, removes “ghost” cells, and yields higher-quality data for downstream analyses
InSituDiff: Disease-Driven Changes in Cellular Neighborhoods as a Window into Spatial Transcriptomics
This poster introduces InSituDiff, a new computational framework for identifying how disease or other perturbations reshape cellular neighborhoods based on CosMx SMI spatial transcriptomics data. By comparing neighborhoods in infected or diseased tissues to their closest match in healthy controls, InSituDiff builds a “perturbation matrix” for each gene, highlighting which genes (and which areas) are most changed. We then employ standard single-cell analytics (clustering, network analysis, etc.) on these perturbation profiles, revealing spatially correlated gene modules and distinct “perturbation domains.”
Connecting Form and Function: Mapping Microglial Spatial Biology in a Mouse Model of Acute and Chronic Ischemic Stroke
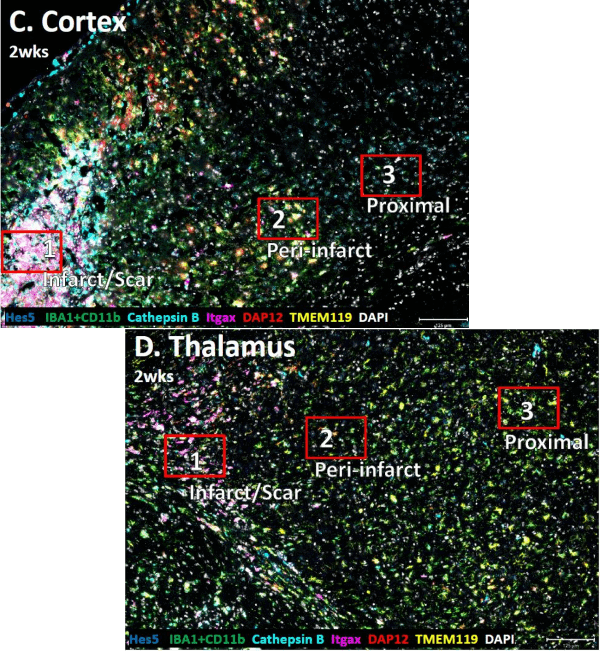
This poster highlights how CosMx SMI can map microglial form and function in the mouse brain following ischemic stroke. By combining single-cell proteomics with morphological analysis, our collaborative partnership with University of Arizona researchers captures how microglia transition from surveillance to injury-response states and reveals distinct gene expression patterns in infarcted versus healthy regions. Notably, we demonstrate that proteins like TMEM119, Cathepsin B, and DAP12 change in both intensity and spatial distribution as you move away from the infarct core, indicating functional heterogeneity across brain regions.
BONUS WEBINAR Expanding the Limits of Spatial Biology: Comprehensive Integrated Technologies for Spatial Multiomics
Our Silver Sponsor Workshop is now available to stream on demand! Catch up on all of our technology announcements:
- 3D genome exploration with the new PaintScape™ platform
- Pathways-first biology with the world’s only unbiased, single-cell whole transcriptome solution using CosMx SMI
- Unparalleled discovery multiomics with GeoMx
® WTA + over 1000 proteins - Improvements in data and image quality with the PowerOMX™ engine for the CellScape platform
- New customer success programs


Related Content