How Do I Choose the Right Spatial Biology Technology?
What is Spatial Biology?
The rapidly evolving field of spatial biology investigates the spatial location and organization of gene expression in situ within each cell and structure of a given tissue sample. Maintaining the spatial context of biological data is crucial for understanding how cells organize and interact with their surrounding environment to drive various biological functions.
Thus, spatial transcriptomics and spatial proteomics can be used to study both healthy and diseased tissues by understanding the mechanisms behind the development, tissue heterogeneity, and therapeutic response. Historically, spatial patterns of gene expression were captured by immunohistochemistry (IHC) and in situ hybridization (ISH). These techniques provided researchers with the ability to visualize biological processes spatially by directly mapping the location of DNA, RNA, and protein molecules within tissues. However, these methods limited analysis to at most a handful of genes or proteins at a time.
The Evolution of Spatial Biology
A breakthrough for transcriptomics in the late 2000s was bulk genomic sequencing which replaced microarray technology. Bulk RNA sequencing involved sequencing of complementary DNA (cDNA), in which abundance is derived from the number of counts from each transcript.
Although bulk RNA-seq is high-throughput and can quantify expression levels of numerous genes across a large population of cells, it obscures biologically relevant differences between cells, and spatial context is lost. Part of this problem was resolved with the advent of single-cell RNA-sequencing (scRNA-seq), which allows for transcriptional profiling of individual cells instead of a population of cells1.
However, both bulk RNA-seq and scRNA-seq still fail to preserve the spatial organization of cells and their associated transcripts within the tissue. Parallelly, the advent of Next Generation Sequencing (NGS) enabled parallel multiplexed analysis of DNA sequences on a massive scale—tens of thousands of sequences from individual single strands of DNA could be analyzed separately yet simultaneously.
Spatial Transcriptomics Technologies
Biology that is inherently spatial—such as cell fate decisions that depend on spatial relationships between cells or the spatial interaction of immune cells within a tumor—cannot always be resolved by bulk analysis or single-cell imaging. Several new technologies have been developed that aim to combine gene expression data with spatial context.
NGS-Based vs. Imaging-Based
Most of the spatial transcriptomics technologies available today have been built on in situ and scRNA-seq and can be primarily categorized into (1) NGS-based approaches that encode positional information onto transcripts before sequencing and (2) imaging approaches based either on in situ sequencing (ISS) where transcripts are amplified and sequenced in the tissue or fluorescence in situ hybridization (FISH) where imaging probes are sequentially hybridized in the tissue2. This classification is not clear-cut as some technologies may incorporate elements of both categories.
Which Spatial Transcriptomics Method Should You Choose?
While both NGS- and imaging-based technologies are extremely powerful, each has its limitations. No single method can currently address all the desired parameters such as high throughput, sensitivity, resolution, and sample compatibility.
High-throughput NGS-based methods are unbiased as they capture all polyadenylated (polyA) transcripts making them better suited for true discovery applications or scanning an entire tissue sample. However, with these methods that rely on a polyA pulldown, there is always the risk of missing lower abundance transcripts without compensation with respect to sequencing depth.

Digital Spatial Profiling
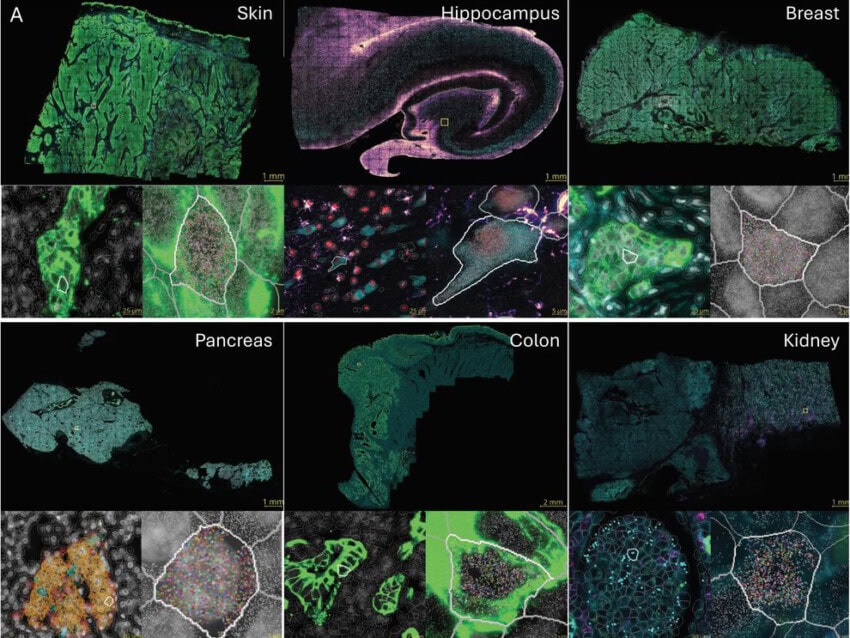
Targeted NGS-based methods like Digital Spatial Profiling (DSP) used by the GeoMx® DSP System allow researchers to profile up to 18,000+ protein-coding genes—the whole transcriptome—while maintaining a wide dynamic range for detection of low to high expressing genes. GeoMx DSP works well on notoriously difficult Formalin-Fixed, Paraffin-Embedded (FFPE) tissue sections and to date has been a valuable tool for research in oncology, COVID-19, developmental biology, and neuroscience. FFPE tissue sections that have been collected over decades and remain untouched sitting in biobanks can now be used for retrospective transcriptome analyses involving not only biomarker discovery but atlasing of the whole transcriptome in different tissue substructures.

Single-Cell Imaging
Targeted and imaging-based spatial transcriptomics, on the other hand, are best suited when both single-cell resolution and subcellular transcript localization are required. However, imaging-based approaches are limited in terms of throughput.
In situ sequencing approaches directly read out the transcript sequence within a cell; although the sensitivity of this approach is high, the resolution is limited by optical diffraction. Multiplexed FISH methods provide the highest gene detection sensitivity but are disadvantaged by molecular crowding when scaling up the plex of the number of genes.
One of the emerging targeted and imaging-based technologies that bridge the gap between high plex, high throughput, and high resolution is NanoString’s CosMx® Spatial Molecular Imager (SMI). CosMx SMI is a multiplex in situ-based system that provides single-cell and sub-cellular resolution on FFPE and fresh frozen tissue sections with quantification of up to 1000 RNAs or up to 100 proteins. Because CosMx utilizes smart cyclic in situ hybridization chemistry with an “on” and “off” signal barcode design that overcomes molecular crowding, only a subset of RNAs or proteins is “on” in any given cycle, leaving room for continued expansion of more plex.
With a tunable workflow from high-plex to high-throughput, CosMx enables not only the highest plex and unbiased whole-slide analysis but also low-plex, high-throughput biology-driven analysis. The applications of CosMx are vast and versatile: it could be utilized to discover and map cell types, generate cell atlases, analyze cell-cell interactions, and discover single-cell biomarkers.
Digital Spatial Profiling Meets Single-Cell Imaging
The CosMx SMI system is ideal for deeper single-cell exploration downstream of whole transcriptome analysis carried out on GeoMx DSP. These two systems can essentially be used for complementary experiments on a set of samples. As single-cell resolution spatial transcriptomics and proteomics pioneer the next frontier in spatial biology, our understanding of fundamental processes in biology will deepen and perhaps pave the way for next-generation diagnostic and pathology tools3.
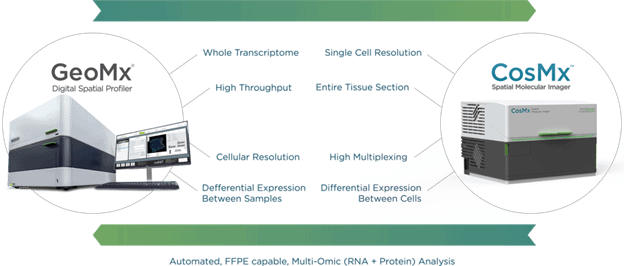
GeoMx DSP and CosMx SMI together are the answer to our initial question, as they enable spatial multi-omics research at any scale.
References
- Antoine-Emmanuel Saliba, Alexander J. Westermann, Stanislaw A. Gorski, Jörg Vogel, Single-cell RNA-seq: advances and future challenges, Nucleic Acids Research. 2014;42(14):8845–8860
- Rao A, Barkley D, França GS, Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596(7871):211-220.
- Aldridge S, Teichmann SA. Single cell transcriptomics comes of age. Nat Commun. 2020;11(1):4307. Published 2020 Aug 27. doi:10.1038/s41467-020-18158-5
For Research Use Only. Not for use in diagnostic procedures.
The CosMx® SMI and decoder probes are not offered and/or delivered to the Federal Republic of Germany for use in the Federal Republic of Germany for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof in a method used in fluorescence in situ hybridization for detecting a plurality of analytes in a sample without the consent of the President and Fellows of Harvard College (Harvard Corporation) as owner of the German part of EP 2 794 928 B1. The use for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof is prohibited without the consent of the President and Fellows of Harvard College (Harvard Corporation).

Related Content