Standing Out in Space: Tips for Winning a Spatial Biology Grant
Nature Methods is a leading journal for molecular biologists interested in the latest and greatest life sciences research techniques. Each year since 2007, Nature Methods chooses a method to feature that has the potential to make a substantial impact on our understanding of biology. In 2020, the team at Nature Methods chose spatially resolved transcriptomics as the method of the year for 2021.
Spatially resolved transcriptomics or spatial biology fuses two common approaches in molecular biology, microscopy, and next-generation sequencing, to provide unparalleled insight into the spatial context of cells. We can understand the behavior of individual cells and how these cells interface in their social context within tissues. This is especially important in fields like developmental biology, neurobiology, and cancer biology, where cellular location and context are closely tied to cellular behavior and function. It’s truly an exciting time for molecular biology.
What questions do you want to ask in your own research context about cellular biology? What technologies exist to help you ask those questions? How can you bring these technologies to your laboratory?
In this blog article, I provide nine key points about spatial biology, including what it is, how you can use technologies like GeoMx® Digital Spatial Profiler, and strategies for requesting funds for the purchase of GeoMx Digital Spatial Profiler. I’ll also include some ready-to-use resources at the end, including a sample query letter, for you to personalize and use.
Why submit a spatial biology grant?
In the classic children’s story about Goldilocks and the three bears, Goldilocks enters the bear’s house and tries everyone’s porridge, chair, and bed until she finds the one that’s just right. Spatial biology feels just right when we consider how to functionally analyze gene expression.
Cells don’t exist in isolation. Cells communicate with each other and work together within tissues. Looking at gene expression in a whole tissue doesn’t provide enough granularity to really understand tissue dynamics. Although it is helpful for making generalizations and understanding broad patterns, we lose information on rare or unique cell types. It’s “too cold” if we go back to our Goldilocks analogy.
In contrast, cell-specific sequencing provides too fine of a view. Studying gene expression dynamics in rare or unique cells is helpful – but we are missing information about the context. Now we’re “too hot.”
Why is spatial biology just right? Because we’ve captured whole tissue information without losing granularity. Spatial biology provides both context and specificity, facilitating the development of a more precise and contextual understanding of biological processes.
So why include spatial biology in your next grant proposal? Smart utilization of a recent Nature method of the year that gives a detailed and novel perspective of biology will help your grant proposal stand out among the crowd. By asking your research questions of interest in a spatial context, you can generate the novel, actionable, high-impact work that many funders are looking to support. How will you answer your spatial biology questions? With the GeoMx Digital Spatial Profiler.
What is the GeoMx Digital Spatial Profiler? (and how does it work?)
The GeoMx Digital Spatial Profiler provides spatial biology insights in a high-throughput and highly reproducible manner. Researchers can simultaneously capture both spatial and/or morphological information and gene or protein expression through a single sample. The power of the GeoMx Digital Spatial Profiler lies in its ability to provide information on how individual cells function, how cells function in a specific microenvironment, and how cells can function as a unit.
Researchers can choose to work with fresh frozen samples or FFPE (Formalin-Fixed-Paraffin-Embedded) samples. Either RNA or protein can be studied using either fluorescently labeled RNA probes or antibodies, respectively. After staining, the user directs the GeoMx Digital Spatial Profiler to regions of interest. These may include specific structural features within a tissue or regions where different cell types interface, such as between cancerous and non-cancerous cell types. Once the user has defined the region of interest, ultraviolet light is used to release the molecular tags on the antibody or probe. The user can then collect these tags and send them for analysis via Next Generation Sequencing or using Nanostring’s nCounter® technology.
What can I study using GeoMx Digital Spatial Profiler?
One of the overarching fundamental principles of molecular biology is that structure dictates function. If proteins lose their three-dimensional structure, they also lose their function. The single-stranded nature of RNA plus the extra hydroxyl group at the two prime carbon explain why RNAs and not DNAs can have catalytic activity. Hydrogen bonding between adjacent strands of polysaccharides is why cellulose is an excellent structural macromolecule.
Molecular biology principles underlie a wide range of biology subdisciplines, including developmental and cellular biology, structural biology, cancer biology, and neurobiology. As such, there is a broad range of questions that a researcher could ask with the GeoMx Digital Spatial Profiler at their fingertips. Here are some examples:
- How do transcriptional dynamics vary in different parts of a tumor?
- What proteins are expressed in healthy cells immediately adjacent to a tumor?
- How do transcriptional dynamics change as different tissue types develop after gastrulation in a developing embryo?
- How does transcription change temporally during treatment with a drug?
- How does gene expression vary between tissues in healthy individuals? Over the lifespan?
- Are there new biomarkers that scientists could identify that tell us more about an individual’s prognosis after a cancer diagnosis?
- How do disparate cell types in the brain interact with one another?
- How does this interaction change in the context of neurodegenerative diseases, many of which lack effective therapeutics?
I’ve posed some broad overarching questions here and provided some published examples in section four. What questions do you have from within your own specialty?
How have people used GeoMx Digital Spatial Profiler already?
As described in section three, there are a wide range of research questions a molecular biologist (broadly defined) can address using GeoMx Digital Spatial Profiler. When writing a grant to request funds for a capital equipment purchase, reviewers want to know not only what new and exciting questions could be asked,but want proof that the technology has a track record, too. In this section, I’ll highlight some previously published research using GeoMx Digital Spatial Profiler in cancer biology.
As defined by the National Cancer Institute, cancer biology research focuses on the molecular mechanisms underpinning how cells grow (and escape cell-cycle controls to grow at inappropriate times), how cells become cancerous, and how cancerous cells can spread or metastasize through the body. One way the GeoMx Digital Spatial Profiler can contribute to this knowledge is through gene expression analysis in various tumor parts. For example, Dr. Jonathan Chen at the Broad Institute found that IDO1 expression varied between high and low-grade core tumor regions and lymph nodes in patients with colorectal cancer. In another study published in 2019, Trieu My Van and Christian Blank examined transcriptional dynamics between tumor and immune cells in colorectal cancer. Van and Blank found that there was significant antigen presentation at the boundary between the two cell types.
How does GeoMx facilitate collaboration?
Although the stereotypical view of scientists is of an individual with crazy hair working in isolation, collaboration is at the heart of scientific endeavors. Many areas of biological research require multi- and interdisciplinary collaboration. For example, in the context of cancer research, this can require a variety of scientists working together, including biologists, doctors, pathologists, laboratory technicians, and biostatisticians. Many funding agencies specifically look for projects that facilitate inter- and multi-disciplinary collaboration and/or unify multiple institutions on a project.
Coordinating teams, especially disciplinarily diverse teams, is a challenge. However, GeoMx Digital Spatial Profiler’s software facilitates collaboration through its secured remote access capabilities. Anyone across the globe can access data securely through the internet. In addition, principal investigators and/or administrators can assign roles or permissions, facilitating project management.
The GeoMx Digital Spatial Profiler workflow is an asset not only to a research team but is also highly attractive on grant applications and can facilitate faster publication. Faster publication time has a wide variety of benefits. For example, investigators can focus their efforts on research rather than navigating the publication process. For university-based researchers, faster publication times can help students graduate on time and help create a robust publication record for tenure and promotion decisions. This Blog article details a successful collaboration with GeoMx Digital Spatial Profiler, the GeoMx Breast Cancer Consortium.
How does GeoMx promote reproducibility and quality science?
When I was in graduate school, my biostatistics professor discussed issues with statistics and reliability of findings in biomedical research. Many stakeholders are concerned about investigator bias if experiments have appropriate power, and ‘p-hacking’ and the reliability of published results. As a result, researchers have called for increased transparency and focus on the reproducibility of scientific results.
Funders want to know they are choosing projects that will produce good data. Attention to these trends and explaining how your chosen methods promote reproducibility will help your grant be chosen for funding.
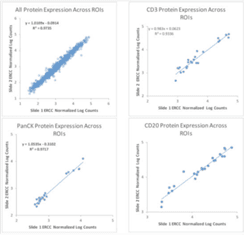
GeoMx Digital Spatial Profiler was designed with this shift in mind. For example, the platform is designed to facilitate collaboration between researchers. As mentioned in section six, one such example of collaboration in action using GeoMx Digital Spatial Profiler is the GeoMx Breast Cancer Consortium. In addition, analysis of serial sections in the tonsil shows the high reproducibility of the results (Figure 1). For additional examples of the reproducibility of GeoMx Digital Spatial Profiler data, see this article by Gupta and colleagues or this Nature Biotechnology article by Merritt and colleagues.
How do I pursue grant funding to purchase GeoMx Digital Spatial Profiler?
Securing grant funding is, unfortunately, a time-consuming and intensive endeavor shared by many scientists. In this Nanostring Blog article, Hiromi Sato outlines some general considerations when applying for grant funding. Keep in mind that several federal agencies offer awards specifically for instrumentation purposes. Depending on the agency, these awards can have different requirements, for example, being shorter than a standard 15-page proposal or having less competition. Depending on which field of biology you hail from, there are also grants specific to your field. For reference, here is a regularly updated and curated list. We’ve also included tips throughout this blog article, but for a quick reference sheet, see section nine for a one-page fact sheet with key bullet points to help guide your grant writing or share with a colleague.
What key methodological details about GeoMx Digital Spatial Profiler should I include in a grant application?
The GeoMx Digital Spatial Profiler not only facilitates spatial biology research but can process over ten sections in a single day, analyzing both RNA and protein. Samples do not require any specialized handling, only standard preservation techniques such as formalin-fixed paraffin-embedded (FFPE) or fresh-frozen samples.
With the next-generation sequencing capabilities, researchers can analyze tens of thousands of distinct targets. Minimum segment size requirements for tissue types are based on cell size and range from one to five cells for large cells like neurons for protein analysis (or 5-20 for RNA analysis) and 10-20 small cells like T cells for protein analysis or 50-200 cells for RNA analysis.
The integrated workflow facilitates project management across diverse teams, regardless of if they are housed within the same building or on the other side of the world. This increases team effectiveness and facilitates a fast process from initial conception and planning to publication.
Where can I find more information?
Selections from the Nanostring Blog:
- How do I choose the right spatial biology technology?
- Spatial Revolution: An exciting future for cancer biology research
- Spatial Transcriptomic Services: Azenta Life Sciences Paves the Way
- The importance of segmentation in spatial biology
- From the Nanostring Blog: Breast Cancer: Best Practices for Using GeoMx Digital Spatial Profiler
- From the Nanostring Blog: Spatial Transcriptomics: Birth of GeoMx Digital Spatial Profiler
- From the Nanostring Blog: Why Spatial Biology?
For Research Use Only. Not for use in diagnostic procedures.

Related Content