Spatial Transcriptomics: Birth of GeoMx® Digital Spatial Profiler
“A rose started off a bud, a bird started off an egg, and a forest started off a seed.” (Matshona Dhliwayo, author and philosopher). It may seem odd, but the story of spatial transcriptomics starts off with a potato.
What is Spatial Transcriptomics
Spatial transcriptomics defines an array of technologies enabling researchers to locate transcripts down to the subcellular level, providing an unbiased map of RNA targets throughout tissue sections. NanoString had the perfect background to pioneer such technology as we specialize in molecular biology tools for translational research.
The NanoString nCounter® System allows one to profile hundreds of RNA, DNA, or protein targets using single-molecule imaging of molecular “barcodes”, enabling digital quantitation of hundreds of unique targets in a single reaction. Such a technology delivers robust data on all samples, including formalin-fixed paraffin-embedded (FFPE) tissue.
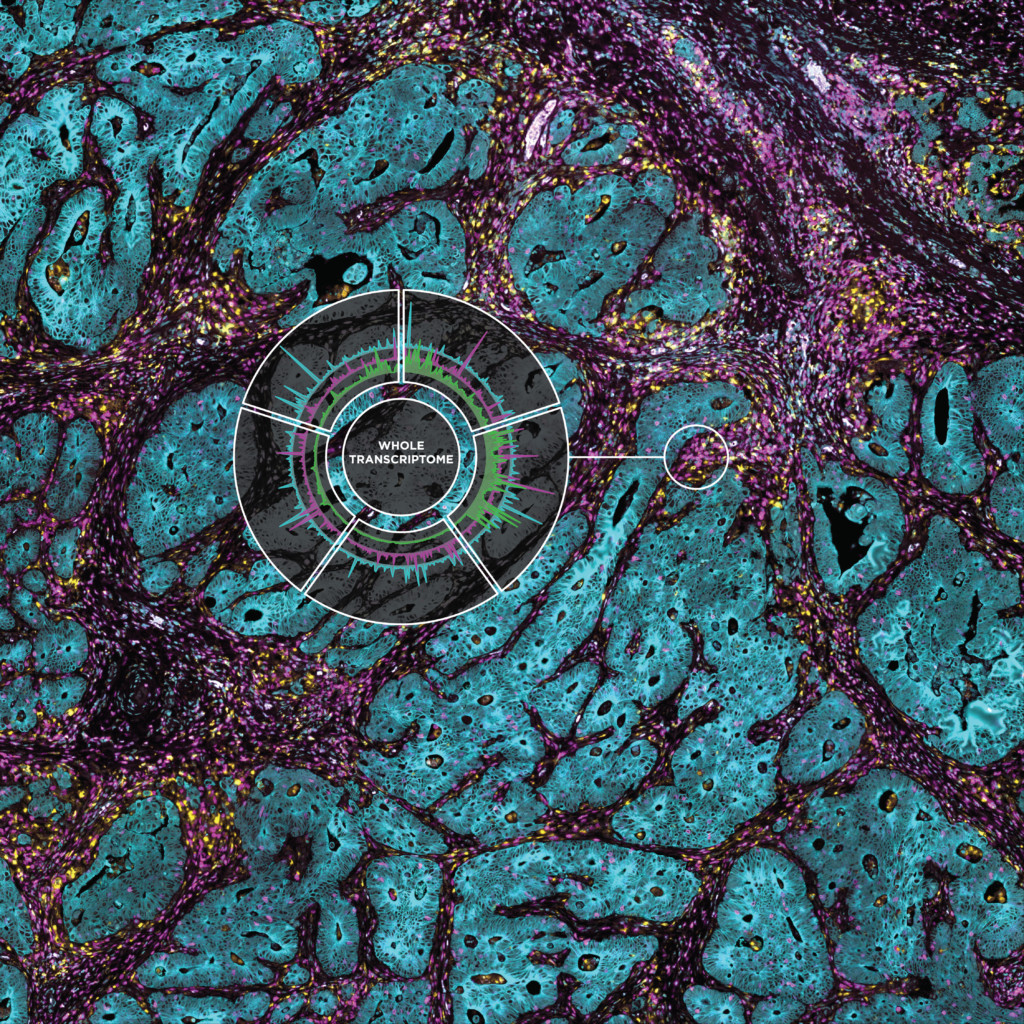
Evolution of Spatial Transcriptomics
Our technology started with mRNA target detection and soon grew to include DNA and miRNA. In 2014, we received the request that would change everything and make NanoString the pioneer in spatial transcriptomics. Ralph Weissleder from the Massachusetts General Hospital wanted to perform multiplex protein detection on clinical tissues.
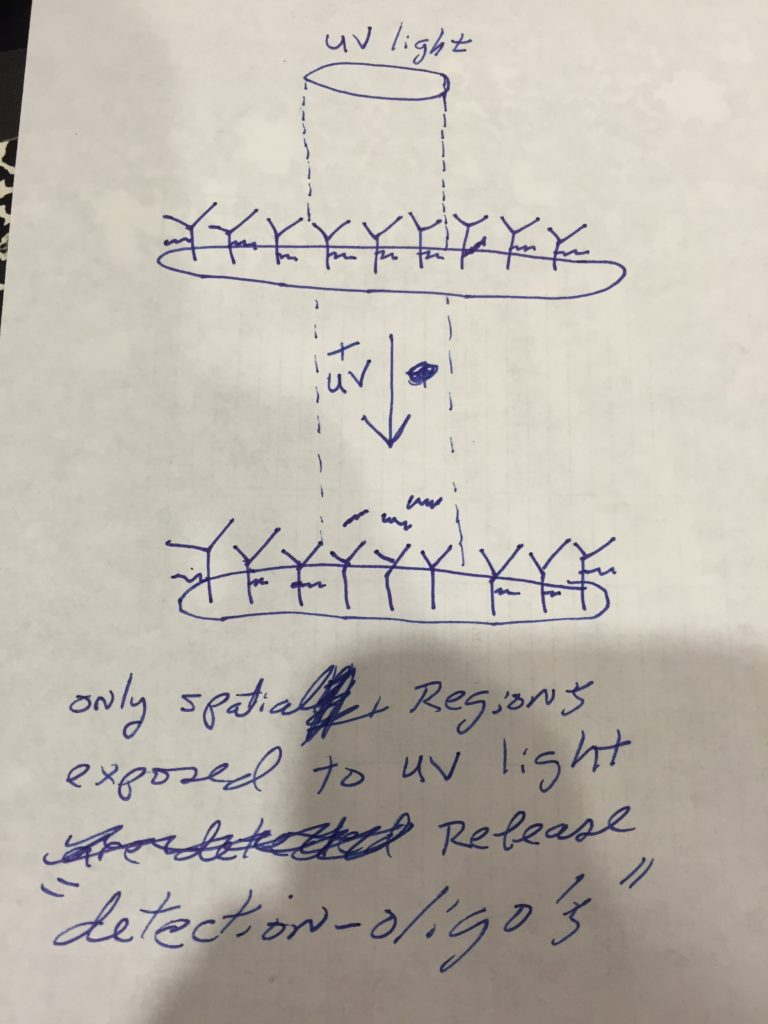
Dr. Weissleder’s team had the idea of adding a unique DNA barcoding to each antibody with a photocleavable linker: once the antibody was bound to the cells, the photocleavable linker would release the unique DNA barcode, which would then be detected and counted.
A team at NanoString led by Joe Beechem, our Chief Scientific Officer, was testing a number of protein assay possibilities with oligo-labeled antibodies, therefore they were already equipped for the challenge. With Joe’s leadership, they conjugated antibodies to DNA sequences derived from the potato genome (easy to make non-reactive with mammalian tissues) and matched them to unique oligonucleotide barcodes to enable direct quantification of proteins on the nCounter platform. The project was successful, with 82 proteins tagged using these potato sequences on antibodies, which led to a publication in Science Translational Medicine.
This technology eventually led to some NanoString product launches that were called “3D-Biology”, but the absolute protein sensitivity wasn’t what R&D really aspired to accomplish. We were not quite ready yet to enter the world of spatial transcriptomics.
Spatial Resolution on a Napkin
The answer came in Houston, Texas, right before a conference where Joe was scheduled to give a talk. He was in his room at the Marriott Hotel, waiting to walk across the street to the MD Anderson Cancer Center when he started thinking: what can we do with antibodies labeled with photocleavable barcoded antibodies that would increase the sensitivity to protein detection that are also spatially resolved? Can we build a microscope or an imaging system to visualize the antibody locations and then use the photocleavable linker to “spatially localize things”? He jotted the idea down on a hotel pad and left to go give his talk.
Later that evening, Joe was with Gordon Mills, then chair of the Department of Systems Biology at MD Anderson, when he realized they both were thinking of very similar ideas. The two started brainstorming, drawing on a whiteboard to scope out if-and-how such a system would work, and what making such technology would require, all starting from the memo pad Joe had in his pocket.
This Could Work!
Before leaving for the evening, Joe took a snapshot of the whiteboard. All of this was accomplished in person and in emails between January 6th and January 13th, 2015. Waking up the day after, Joe looked at the snapshot of the whiteboard and at the memo pad, telling himself: “what were we thinking last night?” but the more he looked at the image, the more he realized: this could actually work!
Back in Seattle, he proposed this new idea to his team. As with any new idea, scientists tend to be a “skeptical group,” but one of the scientists started working on a proof of concept at night so that it wouldn’t interfere with his “day projects.”
He stained microscope slides with the antibodies attached to the photocleavable linkers, using tape on half of the bottom of the slide to block UV light as a control. When the UV light hit the slide, the signal was extremely high – 20 times higher than what they were expecting – and very little noise. With the principle proven to work as expected, the project shifted from a one-person to a three- to four-person project, and the team that created GeoMx began to take shape, kicking off the era of spatial transcriptomics.
Pioneers in Spatial Transcriptomics and Spatial Biology
The next challenge was to convince the public – and the investors – that a technology that could visualize a tissue and hundreds of proteins (and eventually 22000 RNAs) with clear spatial resolution was something they needed. Market research analysts were hired to gather data on the market possibilities of such technology.
One problem: there was no market for a technology that didn’t (yet) exist. Therefore, the market predictions for the commercial return for this product were grim and, at least according to the firm, spatial biology was not viable. On top of that, a few pathologists, when Joe showed them some preliminary data, discarded the idea of high-plex spatial biology as too overwhelming: already 3 to 4 proteins on a slide are hard to interpret, imagine thousands!
Joe, undaunted, took this information back to an investor meeting but also presented sound preliminary data and included the concept of NanoString as the pioneer of spatial transcriptomics. After much preliminary data, and much championing (an essential component of developing any transformative technology), the project was greenlighted and funded.
On March 27, 2019, at the AACR meeting in Atlanta, GA, the GeoMx Digital Spatial Profiling platform was officially launched, with ~ 80-plex protein panels and ~ 100-plex RNA panels, with a focus on Immuno-Oncology applications. During the next two years, the RNA-plex would advance to ~ 2000-plex (in 2020) to whole transcriptome for both human and mouse (~ 22000-plex) in 2021. The protein-readout also advanced to utilize NGS-readout (identical to high-plex RNA), yielding unlimited protein-plex, with currently over 300 validated NGS antibodies.
Where we are today
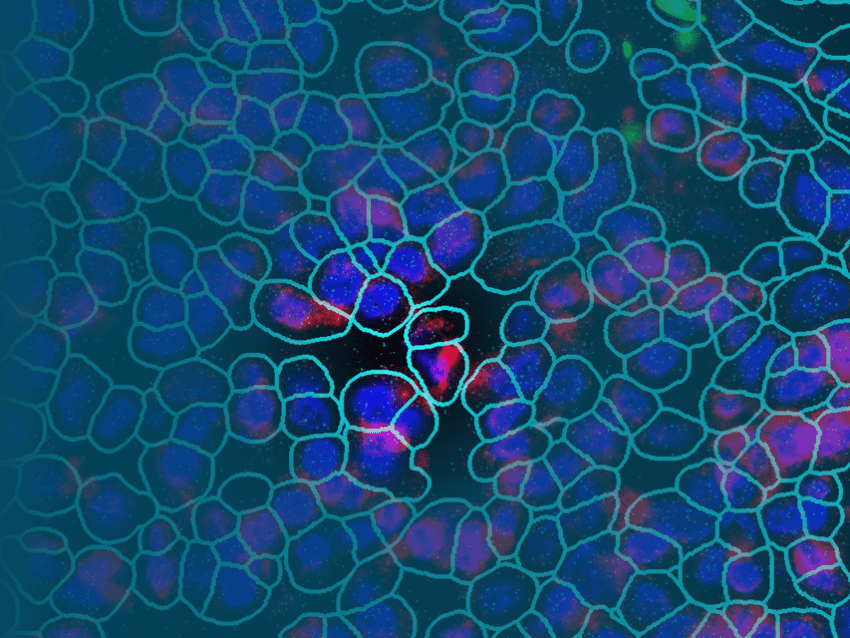
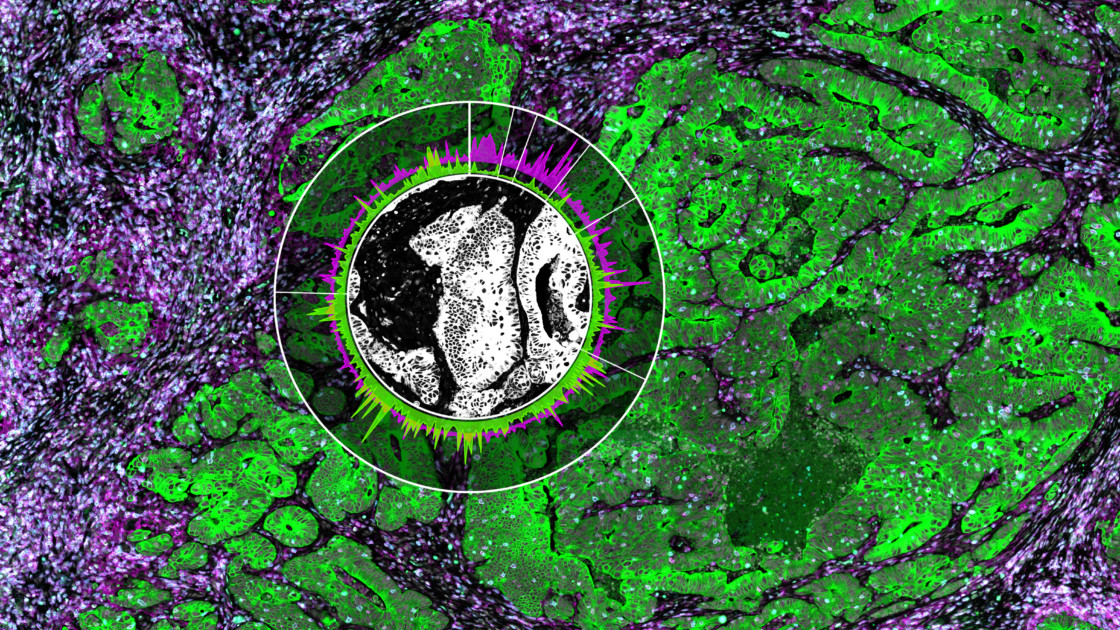
The GeoMx Digital Spatial Profiler is the only fully integrated platform capable of spatial resolution of both RNA and Protein detection on the notoriously tricky FFPE sample type (all sample types work with GeoMx). Both proteins and RNAs are attached to oligonucleotide tags bound to affinity reagents (antibodies or RNA probes) through a photocleavable (PC) linker. The photocleaving light projected onto the tissue sample releases the PC oligonucleotides in any spatial pattern across a region of interest covering 1 to thousands of cells, exactly how Gordon Mills and Joe Beechem “drew-it-up” on a napkin in 2015. GeoMx enables RNA and protein for both nCounter and Next-generation Sequencing readouts.
In January 2021 spatial transcriptomics was nominated 2020 method of the year by Nature Methods.
And 2021 is far from over…and just remember, it all started with a potato and a napkin!
FOR RESEARCH USE ONLY. Not for use in diagnostic procedures.

Related Content