A new map for our anatomy: the Spatial Organ Atlas
One year for spring break, we found ourselves in Portland, Oregon. For science-y people, a trip to Portland would be incomplete without a visit to the Oregon Museum of Science and Industry or OMSI. The oldest exhibit at OMSI lives in the natural sciences hall – a glass woman. She came to the museum in 1958. Her counterpart, the glass man arrived in the United States in 1933 to be displayed at the World’s Fair in Chicago. OMSI’s glass woman is currently one of five still on display in the United States.
The glass woman is a life-size transparent woman. Visitors to the museum can press various buttons which light up her organ systems. In the process, learning about the location and function of various organ systems.
Understanding how normal organs function is important because it gives insights into how things can go wrong. For example, improving our knowledge of how diseases develop. If we know how both normal and diseased organs function, we can devise better prevention strategies and treatments.
Although the glass woman demonstrates how we can understand the basic structure and location of organs, we’re still missing an important piece of the puzzle: the cellular and molecular anatomy. How does gene expression change not only between organ systems but within space?
NanoString® Spatial Organ Atlas attempts to do just that. In this blog article, I’ll introduce spatial biology and the need for a spatial organ atlas. I’ll then discuss how GeoMx Whole Transcriptome Atlas and GeoMx® Digital Spatial Profiler were used to characterize spatially resolved transcriptomics in both the kidney and pancreas. Finally, I’ll close with how researchers can leverage GeoMx Whole Transcriptome Atlas and GeoMx Digital Spatial Profiler in different applications.
Why spatial biology and a Spatial Organ Atlas?
In this article on cancer biology research, I liken spatial biology to the children’s game, hide and seek. Typically hide and seek occurs on a 2D plane. You can hide in the closet or in the bathtub. When we think about the history of spatial biology, this is like in the mid-2000s when genomic analysis meant bulk sequencing, with no information on spatial sorting. It yielded important information but didn’t give the whole picture.
By 2012 researchers were sequencing individual cells. It was like finding that extra special hiding spot in hide and seek. Single-cell sequencing facilitated our understanding of the biology of single cells, and in particular rare or unique cell types. But we were still missing the 3D context. We were still limited to a single aspect of the study- just like the hide and seek game that was limited to the 2D plane.
What if hide and seek involved 3D space? It becomes much more difficult to find someone if they can hide in the branches of a tree. The game reaches a new level.
We see a similar occurrence with cell biology as well. Spatial context is a critical part of cell biology and important for understanding everything from cancer to developmental biology. In 2019, scientists were able to look at cells in their spatial context for the first time using GeoMx Digital Spatial Profiler. Now we can view cells not only as individual entities but as part of their dynamic spatial context within organs. Understanding the molecular anatomy of tissues will help scientists gain a new view on organ function and physiology.
NanoString’s goal in generating a Spatial Organ Atlas was to supply researchers with a benchmark of transcriptional dynamics within certain functional spaces within organs (for example, the basic filtration units of the kidney, also known as glomeruli. These baseline measurements can then be used by other scientists who, for example, want to compare a diseased organ to the standard organ. Given the importance of structure driving function in biology, researchers at Nanostring chose key functional units of five different organs, kidneys, pancreas, brain, colon, and lymph nodes for their pilot studies.
Characterization of transcriptomics in the kidney and pancreas
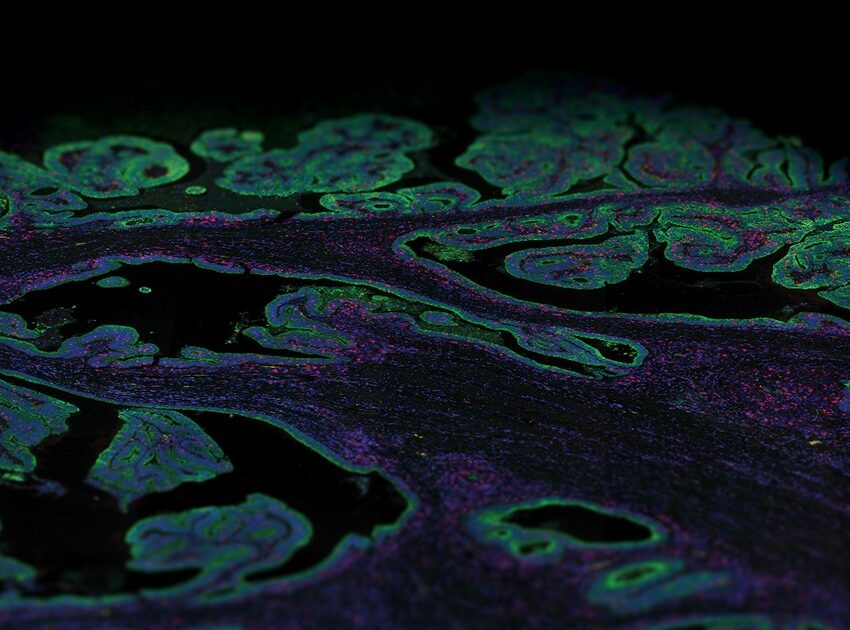
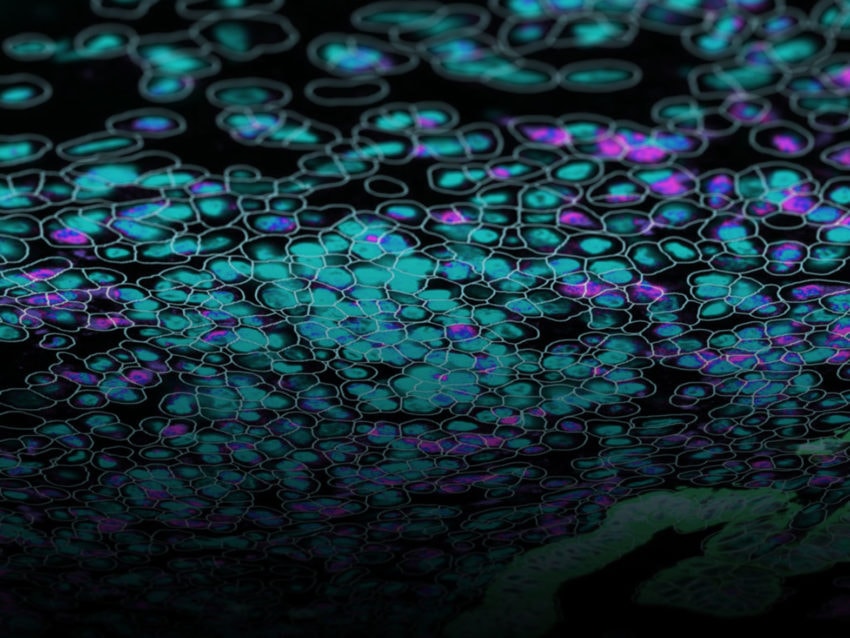
To characterize transcriptomics in organs, first, scientists used the GeoMx Whole Transcriptome Atlas Assay to detect all of the protein-coding genes in a tissue. Next, pathologists looked for particular morphologies that are hallmarks of key structural components of the organs to select areas of interest for analysis with the GeoMx Digital Spatial Profiler. The researchers then used ultraviolet light to cause molecular tags to be released within that region, which were then collected and processed using Next-Generation Sequencing.
There are a wide variety of diseases that impact the kidneys. The kidney is important for filtering the blood, and loss of kidney function can lead to the need for either dialysis or a kidney transplant. NanoString Sr. Director of Pathology Yan Liang and colleagues set out to analyze four healthy kidneys for the purpose of demonstrating proof of principle for the Spatial Organ Atlas. They decided to focus on structures within the kidney nephron, the functional unit of a kidney. These functional structures included the glomeruli, the glomerular filtration membrane, the proximal and distal convoluted tubules, the loops of Henle, and the collecting ducts. The research team found spatially differentiated gene expression between regions, such as between the cortical and juxtamedullary glomeruli.
Sr. Bioinformatic Scientist Erin Piazza and colleagues worked to characterize spatially resolved transcripts within the pancreas. The pancreas is an important area of study because dysfunction of the pancreas is related to diabetes, a condition that impacts 1 in 10 Americans. Furthermore, Pancreatic cancer has a combined five-year survival rate of 11% – and its diagnosis is devastating for families.
In their work, Piazza and colleagues analyzed functional structures such as the Islets of Langerhans, pancreatic acini, and ducts from four healthy samples. Piazza and colleagues note in their work that they were able to detect cell-type-specific genes in their appropriate locations, providing evidence that their spatial analysis of functional structures within the pancreas was accurate.
Concluding remarks and other applications of the spatial organ atlas
An atlas is a book of maps – and maps are an essential tool for finding our way around. Maps change overtime too – just like our understanding of anatomy has changed over the past 90 years from a glass woman whose organ structures are mapped out at a gross level, to now where we are refining our maps to include not only information on functional units of organs but the underlying gene expression signatures within those functionally critical units.
Utilization of the GeoMx Whole Transcriptome Atlas alongside the GeoMx Digital Spatial Profiler to generate evidence-based spatial biology benchmarks will be useful across biological disciplines. For infectious disease specialists, how do transcriptional dynamics in an organ change as a factor of viral load? Can this explain organ damage caused by infectious diseases? How does spatial gene expression in the brain change in people with neurodegenerative diseases? How does gene expression change as an organ like the brain develops as children grow up?
What questions will you ask with the Spatial Organ Atlas?
For Research Use Only. Not for use in diagnostic procedures.