Unveiling the Inner Lives of Cells: A Spatial Look at Cell-Cell Interaction
Cells are the building blocks of all living things, from single-celled organisms such as bacteria to multi-celled organisms like whales. At the most basic level, though, all cells function in very similar ways – they harbor genetic information, they metabolize energy sources, they renew themselves, and they sense their environment.
One significant difference between single-celled organisms and multi-celled organisms is the degree to which cells interact with each other. Also known as cell-cell communication, cell-cell interactions between single-celled organisms often occur over some distance by chemical signals, often without physical contact. Multicellular organisms, however, not only utilize long-distance chemical or molecular interactions, but also direct physical interactions. Studying these interactions is key to understanding biological systems on a molecular level.
To date, molecular information about cell-cell interactions has come from several techniques, including yeast two-hybrid screening, co-immunoprecipitation, proximity labelling proteomics, fluorescence resonance energy transfer imaging and X-ray crystallography.
Used to identify many interactions between secreted or extracellularly displayed proteins involved in intercellular communication, traditional techniques are now being enhance by current proteomics and transcriptomics technology to further understand protein-protein interactions.1Armingol E, Officer A, Harismendy O, Lewis NE Deciphering cell–cell interactions and communication from gene expression. Nat RevGen (2021) Vol 22: pp 71-88 To date, communication between over 144 human primary cell types has been characterized. While direct analysis by proteomic technologies is the most straightforward approach for establishing these interactions, characterizing the presence of RNA transcripts is easier and faster while still providing reliable information. Known as transcriptomics, these results need extra analysis to confirm the presence of the associated protein products, requiring techniques capable of colocalizing cognate ligands and receptors in cells.1Armingol E, Officer A, Harismendy O, Lewis NE Deciphering cell–cell interactions and communication from gene expression. Nat RevGen(2021) Vol 22: pp 71-88
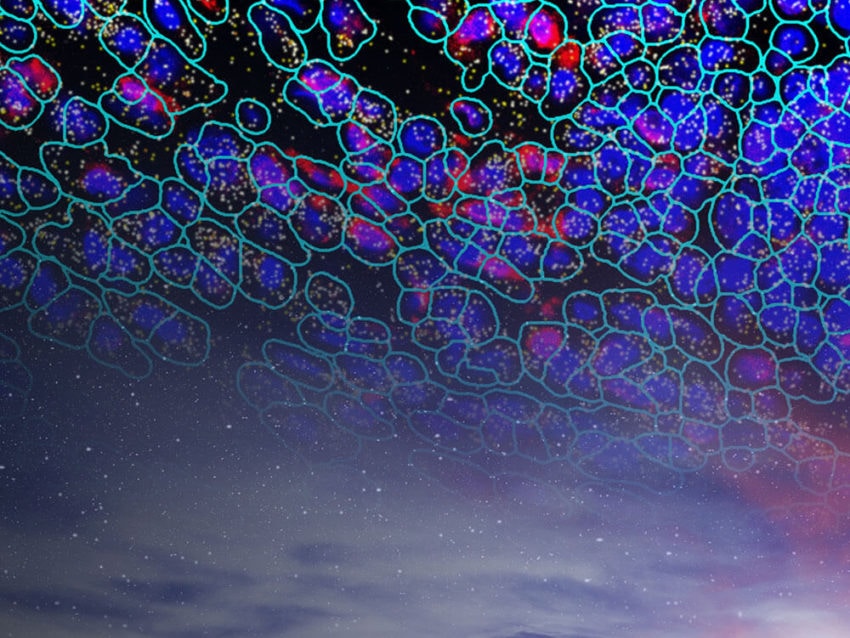
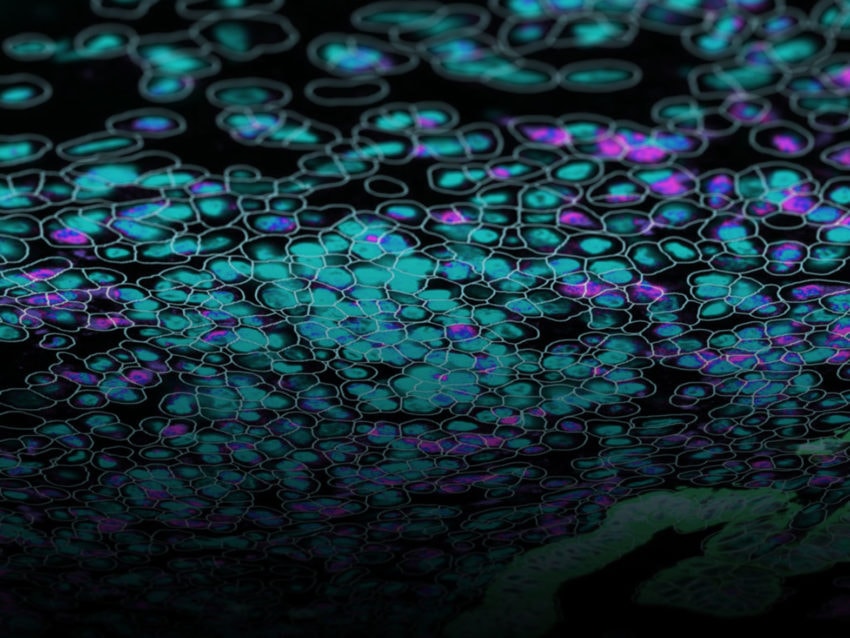
Spatially resolved multiomics using CosMx SMI is facilitating new discoveries into cell-cell interactions, which have implications for diseases such as cancer.
A critical inadequacy of traditional techniques is the inability to simultaneously identify, in a high-plex manner, transcripts and proteins in their native spatial context. Recent technologies harness high power bioinformatic analysis to examine a tissue’s entire transcriptome and proteome on a single slide, directly unraveling both long-distance and direct contact cell-cell interaction. NanoString’s CosMx™ Spatial Molecular Imager (SMI) examines cell-cell interactions by analyzing the multiomics (protein and RNA expression data) of intact tissue, providing spatial context for the expression profiles. In this way, cognate ligand and receptor proteins are simultaneously localized with associated RNA expression in specific cells. Spatially resolved multiomics using CosMx SMI is facilitating new discoveries into cell-cell interactions, which have implications for diseases such as cancer.
What are cell-cell interactions?
A cell-cell interaction is any biological communication between cells.
Cell-cell interactions come in a variety of forms, most of which fall into one of two categories: direct physical contact and non-contact communication. Both categories illustrate a fundamental principle of cellular biology that states proper function relies on appropriate tissue structure along with temporal and spatial coordination of molecular and cellular events.
Collectively maintaining the dynamic equilibrium of an organism, the various types of interactions between cells across tissue space and time are key to understanding complex cellular functions. Knowing more about normal cell-cell interactions can provide insight into disease development processes that occur when specific interactions are disrupted.
What are some ways that cells communicate with one another?
Cell-cell interactions involving direct contact have various functions, with cellular communication being a key one. However, some direct interactions are mainly structural. Tight junctions, for instance, provide a tight barrier between cells, regulating molecular traffic by preventing certain molecules from travelling in the intercellular space. The blood-brain barrier is a great example, consisting of tight junctions between the endothelial cells of brain capillaries, forming a semi-permeable barrier that allows only certain molecules (oxygen, water, a few others) to transfer from the blood to the brain. While serving a mainly barrier function, tight junctions can be involved in cell-cell communication by playing a role in signal transduction pathways.2Takano K, Kojima T, Sawada N, Himi T. Role of tight junctions in signal transduction: an update. EXCLI J. 2014 Oct 13;13:1145-62. PMID: 26417329; PMCID: PMC4464418.
Other direct contact interactions, like gap junctions, facilitate both communication and physical connection between cells by forming a protein-constructed gateway. Gap junctions form a pathway between cells, often regulating various molecules, ions, and electrical impulses to pass from one cell to the other. Gap junctions in the heart help coordinate heartbeats by passing the signal to contract from one cardiac muscle cell to the next.
Ligand-receptor interactions
Since tight junctions and gap junctions form a direct physical connection between neighboring cells, defining the spatial tissue context of the cell types involved is relatively easy. Ligand-receptor interactions, however, typically involve more than one cell type. Further complicating the identification of the cell types and tissue locations involved is the different ways ligands and receptors are expressed and interact. Ligands can act in direct cell-cell contact manner as well as at a distance for non-contact communication. Spatial multiomics can therefore be highly valuable for identifying the cells involved in these interactions.
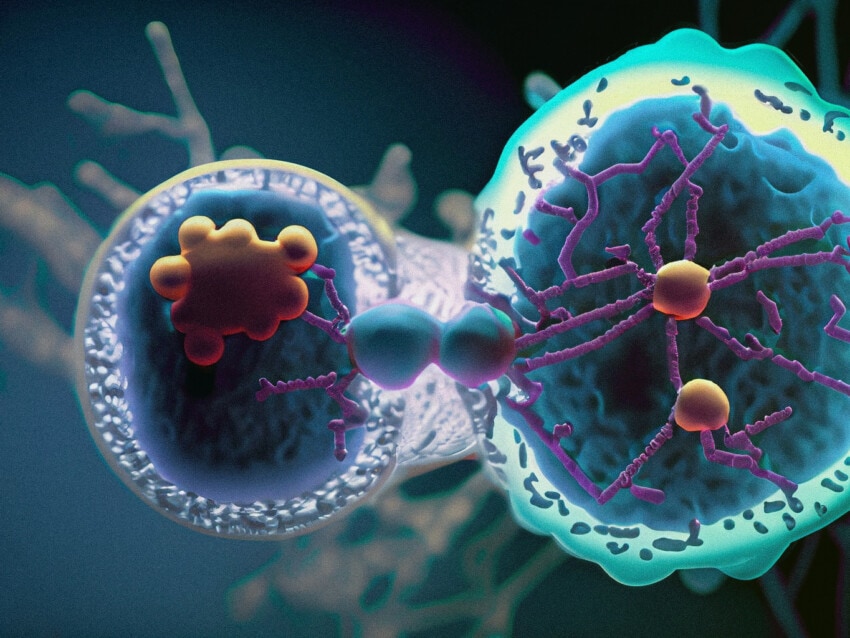
The most straightforward ligand-receptor interactions are between neighboring cells that directly signal each other. Known as juxtacrine signalling,1Armingol E, Officer A, Harismendy O, Lewis NE Deciphering cell–cell interactions and communication from gene expression. Nat RevGen(2021) Vol 22: pp 71-88 a membrane-bound ligand on one cell binds to a membrane-bound receptor on a neighboring cell. While spatially proximal, direct ligand-receptor interactions often involve different cell types that can be differentiated by spatial multiomic technology. A prime example of direct cell-cell interaction via ligand-receptor binding is T Cell Receptor-Major Histocompatability Complex interactions in the immune system.3Kindt TJ, Goldsby RA, Osborne BA, Kuby J (2007). Kuby immunology. Macmillan. pp. 223. ISBN 978-1-4292-0211-4. Retrieved 28 November 2010.
Non-contact cell-cell communication involves a ligand released by a cell that travels some distance to the receptor-expressing cell. Although the archetype of non-contact cell-cell communication is a ligand-expressing cell distal to the target cell (known as endocrine signaling), the target cell can be either a neighbor cell (paracrine signaling) or even the cell itself (autocrine signaling; Armingol). Such variations in distance from ligand origin to target receptor can make it challenging to identify the responsible cell types. Spatial biology, however, can help identify the cell types involved, or at least eliminate nearby cell types from contention as ligand-producing cells, thus directing the search outward toward to hormonal tissues.
While cell-cell communication via physical perturbations is possible (i.e., microtubules affecting gene expression),4Rosette C, Karin M (March 1995). “Cytoskeletal control of gene expression: depolymerization of microtubules activates NF-kappa B”. The Journal of Cell Biology. 128 (6): 1111–9. doi:10.1083/jcb.128.6.1111. PMC 2120413. PMID 7896875 most cellular communication occurs via ligand-receptor signaling. Cognate receptors for a ligand are found in target cells and can be either membrane-bound or intracellular. The family of nuclear receptors, for example, bind to lipid soluble ligands, such as estradiol or cortisol, that easily traverse the cellular membrane and diffuse through the cell until they encounter and bind their cognate receptor. Once bound, nuclear receptors typically act as a transcription factor, permeating the nucleus and binding to gene promoters containing the appropriate gene sequences . Membrane bound receptors, on the other hand, bind their ligands at the cellular membrane and effect changes in target gene expression through signaling cascades, such as the family of G-protein coupled receptors.5Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009 May 21;459(7245):356-63. doi: 10.1038/nature08144. PMID: 19458711; PMCID: PMC3967846.
Identifying the spatial context of the source cells for the ligand and the target receptor cells is critical to understanding the biology of these cell-cell interactions.
Whatever signaling cascade is used by ligand-bound receptors, the result is signal transmission from outside the cell to inside the cell to effect a change, usually in gene expression. Since the cells that are the source of the ligand instigate the signaling action, a full understanding of the mechanism of a specific signal requires identifying those cells. In addition, identifying the target cells, especially when the ligand is generated by a distant cell type, is necessary for completing the picture of the cell signal cascade. Identifying the spatial context of the source cells for the ligand and the target receptor cells is therefore critical to understanding the biology of these cell-cell interactions.
What is the relationship between cell-cell interactions and human disease?
Since cell-cell interactions are fundamental to biological function, understanding in detail the interactions of cells involved in a biological process establishes a baseline for elucidating mechanisms causing disease. Dynamic biological processes, including cellular differentiation and organ development, tissue homeostasis, and immune system function, rely on cell-cell interactions.1Armingol E, Officer A, Harismendy O, Lewis NE Deciphering cell–cell interactions and communication from gene expression. Nat RevGen(2021) Vol 22: pp 71-88
Cell-cell interactions, in the form of precise cellular communication, are key to proper cellular differentiation. Dependent on the appropriate temporal and spatial expression of the correct signals, mapping the fates of stem cells through differentiation and development has increased based on studies of intercellular communication that revealed ligand–receptor interactions critical for cellular self-renewal and differentiation.1Armingol E, Officer A, Harismendy O, Lewis NE Deciphering cell–cell interactions and communication from gene expression. Nat RevGen(2021) Vol 22: pp 71-88 Tissue spatial context analysis revealed that, due to “the promiscuity of receptors,” the extent of ligand signaling is limited in tissues by physical compartmentalization of cells, conferring specificity of interactions on particular stem cells to guide their differentiation fate.1Armingol E, Officer A, Harismendy O, Lewis NE Deciphering cell–cell interactions and communication from gene expression. Nat RevGen(2021) Vol 22: pp 71-88 Cell-cell communication studies have also expanded understanding of differentiation during epidermal regeneration, development of the interfollicular epidermis and formation of the maternal–fetal interface in early pregnancy.
Tissue homeostasis relies on cell-cell interactions.
Tissue homeostasis also relies on cell-cell interactions. Gene expression profiling can show the dynamics of intercellular communication between different cell types in adult tissues, and analyzing the spatial context of gene expression profiles clarifies the cell-cell interactions across tissues and organs. Measurement of cell-cell interactions by transcriptomic spatial mapping of the responsible cells has aided the identification of a new population of lung endothelial cells (Car4 high) that respond to lung tissue injury.1Armingol E, Officer A, Harismendy O, Lewis NE Deciphering cell–cell interactions and communication from gene expression. Nat RevGen(2021) Vol 22: pp 71-88
Spatial expression analysis has also been used to explore the communication of T cells in the human thymus during their development. A spatially coordinated process, the cellular localization of T cell development was crucial for understanding the mechanism more fully. Localization of cells involved in cell signaling cascades can help elucidate interactions between spatially proximal regions.1Armingol E, Officer A, Harismendy O, Lewis NE Deciphering cell–cell interactions and communication from gene expression. Nat RevGen(2021) Vol 22: pp 71-88
While cellular differentiation and homeostasis are complex biological processes that can lead to disease states, the immune system represents a significant increase in complexity by not only fighting disease as its normal function, but also by causing disease itself when molecular disruptions cause it to malfunction. Identifying the cell-cell interactions of normal immune cells in action and of malfunctioning immune cells therefore requires powerful tools.
Cell-cell interactions involved in coordinating a healthy immune response require highly specific signals to differentiate from the continuous flow of signals received from multiple tissues. The use of single-cell transcriptomes identified the involvement of CCL2- and CX3CL1 as molecular coordinators of immune cell recruitment and positioning during crucial processes combatting ascending uropathogenic Escherichia coli infection.1Armingol E, Officer A, Harismendy O, Lewis NE Deciphering cell–cell interactions and communication from gene expression. Nat RevGen(2021) Vol 22: pp 71-88 However, disruption of cell-cell interactions between immune cells and their environment can lead to a disease state such as leukemia.
Cell-cell interactions involved in coordinating a healthy immune response require highly specific signals to differentiate from the continuous flow of signals received from multiple tissues.
Further complicating the picture of cell-cell communication in the immune system is when local immune cell functions, while still operating as normal, are modified by tumors and the tumor microenvironment (TME) to neutralize the effects of immune cells on the tumor. Understanding the cell-cell interactions within the complex community of cells that compose the TME reveals how cells communicate in these ecosystems, not only guiding the development of effective cancer immunotherapies, but also completing the picture of immune cell interactions in normal disease-fighting processes and in diseased immune systems.1Armingol E, Officer A, Harismendy O, Lewis NE Deciphering cell–cell interactions and communication from gene expression. Nat RevGen(2021) Vol 22: pp 71-88
What is spatial multiomics and what can it tell us about cell-cell interactions?
Since cell-cell interactions are crucial for cellular communication and overall maintenance of biological systems, identifying the who, what, where, and when of the cells interacting with each other is critical to understanding the cellular and molecular biology of a system. Spatial multiomics is a powerful tool for examining the transcriptomics and proteomics of a tissue of interest.
Spatial multiomics examines gene expression profiles, at either the RNA or protein level, in the context of the position of the expressing cells in relation to each other as well as within the context of the tissue. Spatial orientation as well as the biological impact on genomics of cell-cellinteractions is important for understanding not only typical biological functioning but also disease processes. Spatial multiomics preserves information by examining gene expression profiles in intact tissue samples, preserving cells in their biological context.

With high-plex, high throughput capabilities of current technologies such as CosMx SMI and GeoMx® Digital Spatial Profiler from NanoString, entire transcriptomes (and up to 150 proteins) can be analyzed in their spatial context on a single tissue section. These technologies elucidate cell-cell interactions by visualizing and quantifying gene expression profiles and identifying cell types involved. Cells involved in ligand-receptor relationships, as well as other interactions such as tight junctions and gap junctions are easily identified and characterized. In this way, molecular biology of any system can be easily characterized.
How does CosMx SMI provide information on cell-cell interactions?
Cell-cell interactions are key to the proper function of biological systems, since these interactions are the source of the signals for precision changes in molecular processes. The wide variety of interactions, ranging from cells that are physically touching to ones signaling from a distant tissue, mean that identifying the cell types, their location, and the molecules involved are all crucial pieces of the biological puzzle. The ability to screen the transcriptome and proteome of single cells in a tissue in a high-plex, high-throughput manner greatly accelerates the identification of the cells and molecules involved.
Spatial gene expression profile analysis enables researchers to quickly and efficiently characterize the RNA and proteins present in their tissue of interest.
Spatial gene expression profile analysis, such as can be performed by CosMx SMI, enables researchers to quickly and efficiently characterize the RNA and proteins present in their tissue of interest. Using CosMx SMI’s fluorescent barcode hybridization technology, researchers can, for example, examine specific interactions between T cells and cancer cells.6He, S., Bhatt, R., Brown, C. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-022-01483-z7Moutafi MK, Molero M, Martinez Morilla S, Baena J, Vathiotis IA, Gavrielatou N, Castro-Labrador L, de Garibay GR, Adradas V, Orive D, Valencia K, Calvo A, Montuenga LM, Ponce Aix S, Schalper KA, Herbst RS, Paz-Ares L, Rimm DL, Zugazagoitia J. Spatially resolved proteomic profiling identifies tumor cell CD44 as a biomarker associated with sensitivity to PD-1 axis blockade in advanced non-small-cell lung cancer. J Immunother Cancer. 2022 Aug;10(8):e004757. doi: 10.1136/jitc-2022-004757. PMID: 36002182; PMCID: PMC9413286. Characterizing gene expression differences in these cell types can elucidate profiles distinguishing more aggressive cancers from less aggressive ones, thus guiding treatment options.
Further enhancing their spatial multiomics analysis capabilities, NanoString has recently unveiled a cloud based bioinformatics analysis package, the AtoMx™ Spatial Informatics Platform (SIP), that integrates with CosMx SMI for seamless data acquisition and analysis. Using standard formalin-fixed paraffin embedded (FFPE) or fresh frozen samples, CosMx SMI can analyze 1000 RNAs and 64 proteins simultaneously, enabling the user to easily visualize and identify biologically interesting cells.
Understanding cell-cell interactions is the key to unprecedented insight into biological systems
The myriad cell-cell interactions of a multi-celled organism are necessary for its continued proper function. To fully understand the biology of an organism requires a full picture of the spatial multiomic relationships that underlie these cell-cell interactions. Gene expression profile analysis in the spatial context of intact tissue uncovers the important cell-cell interactions of a tissue, and NanoString’s CosMx SMI is proving to be a valuable tool in the field of spatial biology.
The power to extensively describe RNA and protein expression at the single cell and subcellular level will accelerate researchers’ ability to characterize cell-cell interactions of all types, furthering understanding of disease processes and guiding research toward novel treatments.
Spatial biology analysis is leading basic science and medicine toward an exciting future!
The CosMx™ SMI and decoder probes are not offered and/or delivered to the Federal Republic of Germany for use in the Federal Republic of Germany for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof in a method used in fluorescence in situ hybridization for detecting a plurality of analytes in a sample without the consent of the President and Fellows of Harvard College (Harvard Corporation) as owner of the German part of EP 2 794 928 B1. The use for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof is prohibited without the consent of the President and Fellows of Harvard College (Harvard Corporation).
References
- 1Armingol E, Officer A, Harismendy O, Lewis NE Deciphering cell–cell interactions and communication from gene expression. Nat RevGen(2021) Vol 22: pp 71-88
- 2Takano K, Kojima T, Sawada N, Himi T. Role of tight junctions in signal transduction: an update. EXCLI J. 2014 Oct 13;13:1145-62. PMID: 26417329; PMCID: PMC4464418.
- 3Kindt TJ, Goldsby RA, Osborne BA, Kuby J (2007). Kuby immunology. Macmillan. pp. 223. ISBN 978-1-4292-0211-4. Retrieved 28 November 2010.
- 4Rosette C, Karin M (March 1995). “Cytoskeletal control of gene expression: depolymerization of microtubules activates NF-kappa B”. The Journal of Cell Biology. 128 (6): 1111–9. doi:10.1083/jcb.128.6.1111. PMC 2120413. PMID 7896875
- 5Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009 May 21;459(7245):356-63. doi: 10.1038/nature08144. PMID: 19458711; PMCID: PMC3967846.
- 6He, S., Bhatt, R., Brown, C. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-022-01483-z
- 7Moutafi MK, Molero M, Martinez Morilla S, Baena J, Vathiotis IA, Gavrielatou N, Castro-Labrador L, de Garibay GR, Adradas V, Orive D, Valencia K, Calvo A, Montuenga LM, Ponce Aix S, Schalper KA, Herbst RS, Paz-Ares L, Rimm DL, Zugazagoitia J. Spatially resolved proteomic profiling identifies tumor cell CD44 as a biomarker associated with sensitivity to PD-1 axis blockade in advanced non-small-cell lung cancer. J Immunother Cancer. 2022 Aug;10(8):e004757. doi: 10.1136/jitc-2022-004757. PMID: 36002182; PMCID: PMC9413286.