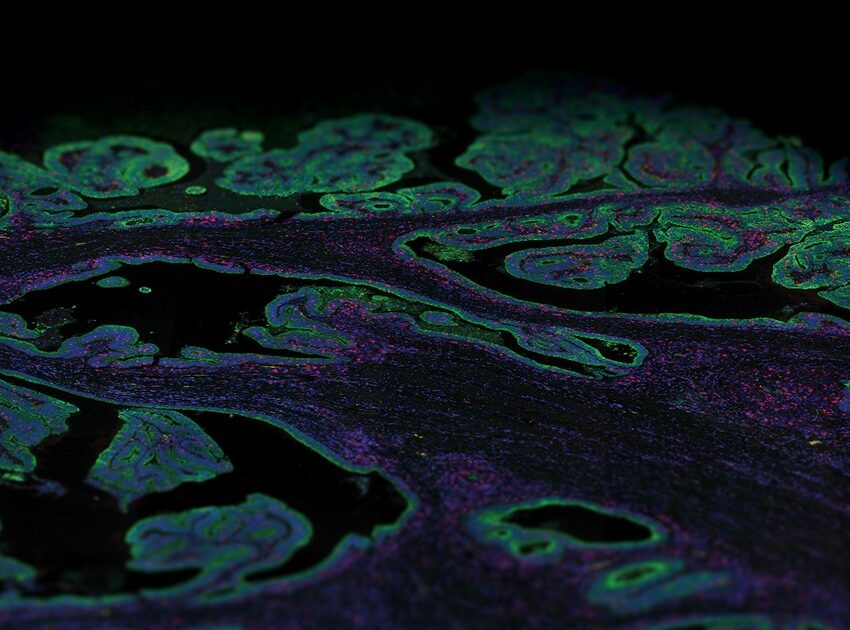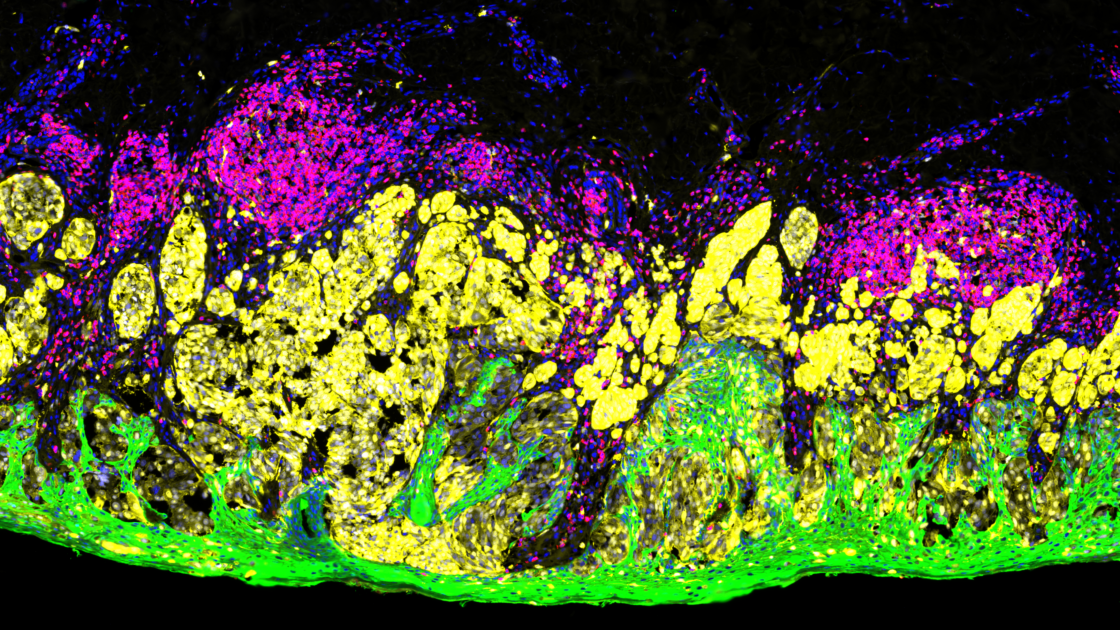
Immunofluorescence Staining: An Overview
Immunofluorescence staining is an incredibly powerful yet simple imaging technique that optically detects the localization, distribution, and abundance of proteins at a cellular and intracellular level using fluorescent tags on fixed biological samples.
Occasionally, terms such as immunocytochemistry (when looking at cells) or immunohistochemistry (when looking at tissues) are also associated with immunofluorescence staining, but these techniques could also use non-fluorescent visualization methods.
Over the past decade, immunofluorescence staining techniques have further evolved to work on FFPE-preserved tissue making it ideal to study the biology of human health and disease thus befitting both basic and clinical research. Immunofluorescence staining has been established as the gold standard for diagnosis of a wide range of tissue pathologies providing valuable information on prognosis, treatment planning, and evaluation of therapeutic outcomes for a variety of diseases.
What is immunofluorescence staining?
Immunofluorescence staining uses fluorophore-conjugated antibodies that make it possible to detect where the target antigen is localized via fluorescence. These fluorophores are excited by one wavelength and emit at a longer specific wavelength that is visible with the aid of a microscope that captures different types of fluorescence as spectral peaks against a dark background. The fluorophores most used for immunofluorescence staining are fluorescein isothiocyanate (FITC) and tetramethylrhodamine isothiocyanate (TRITC). Immunofluorescence is also used in combination with non-antibody fluorescent staining methods, for instance labeling DNA using the reagent DAPI.
Immunofluorescence staining techniques offer several advantages concerning targeting specificity, resolution, signal amplification, and, analytical capabilities, although it is limited to only fixed (dead) cells when structures within the cell are to be visualized. Several other factors also determine the quality and utility of an immunofluorescence technique such as the quality and concentration of the type of antibodies used which must be carefully determined and tested to ensure that a high signal-to-background ratio is achieved.
Types of antibodies and immunofluorescence staining methods
Immunofluorescence staining methods utilize primary antibodies that are either monoclonal or polyclonal which bind non-covalently to the antigen. Immunofluorescence staining methods can be divided into two categories: direct or indirect immunofluorescence. In the direct immunofluorescence method, the fluorophore label is conjugated directly to the primary antibody that binds the target epitope. One of the advantages of direct immunofluorescence is its high specificity and faster turnaround time, but it is limited in sensitivity as there is a limited number of antibodies that can bind the antigen.
Indirect immunofluorescence staining offers high sensitivity, as it allows for six to eight-fold signal amplification.
The indirect immunofluorescence staining method involves a two-step process; first, a primary antibody is used to bind to the target antigen of interest, second, a secondary antibody conjugated to a fluorophore recognizes and binds to the primary antibody at multiple sites. Therefore, this method offers high sensitivity as it allows for six to eight-fold signal amplification.
Amongst the two methods, the indirect approach is more common as most primary antibodies are polyclonal. A major disadvantage of the indirect method is that secondary antibodies can bind endogenous immunoglobulin, introducing background noise.
Multiplex immunofluorescence
Despite the ability of conventional immunofluorescence staining to spatially resolve protein expression, it permits the labeling of only a handful of markers at a time per tissue section. Further, the quantification of protein expression from fluorescent intensity is often not straightforward. Therefore, immunofluorescent methods that can spatially resolve and quantify the expression of many biomarkers at a time are urgently needed to diagnose disease and develop appropriate therapeutic strategies more accurately.
In recent years, the emergence of multiplex immunofluorescence technology that detects multiple proteins in a single tissue section has revolutionized immunofluorescence staining methods. Many of these techniques still rely on antibodies for antigen detection and mapping but have also evolved to simultaneously quantify the detected proteins allowing comprehensive studies of cell composition, functional state, and cell‐cell interactions in the context of the original spatial arrangement of the tissue.
Different types of multiplex immunofluorescence methods
There are various multiplex immunofluorescence methods that have been developed based on five principles: stain removal technologies, fluorophore inactivation technologies, multiplexed signal amplification, mass cytometry, and DNA barcoding technologies. The basic principle of stain removal technologies is repeated staining, imaging, and stain removal; removal occurs with different pH conditions, denaturation, or photobleaching and this method allows for the detection of multiple antigens from a single sample. Multi-epitope ligand cartography is one such automated method that measures several proteins at once from a single sample of cells or tissue and relies on sequential rounds of labeling biomarkers with fluorescent dyes. However, this method is constrained to a single microscope field of view instead of the whole section due to the photobleaching step. The principle of fluorophore inactivation technology is like stain removal technologies but uses chemical inactivation to eliminate the fluorophore. The cyclic immunofluorescence method uses consecutive staining done by iterative cycles of tagging, image scanning, and destaining of a fluorophore on a single slide.
Mass cytometry enables accurate discrimination of metal isotopes of different atomic masses for simultaneous analysis of a much greater number of markers than fluorescence flow cytometry.
Multiplexed signal amplification was developed to detect low abundance protein expression and includes technologies such as multiplex-modified haptens, tyramide signal amplification, and nanocrystal quantum dots. Amongst these technologies, tyramide signal amplification, an enzyme-mediated method that catalyzes the deposition of tyramide on and near a target protein in high density resulting in a high degree of signal amplification, is commonly used. Mass cytometry, a next-generation flow cytometry platform that utilizes elemental mass spectrometry to detect protein expression, was developed to go beyond the multiplexing capability of conventional flow cytometry platforms. Mass cytometry utilizes rare metal isotopes instead of fluorophores for antibody labeling, enabling accurate discrimination of metal isotopes of different atomic masses for simultaneous analysis of a much greater number of markers than fluorescence flow cytometry.
Finally, DNA barcoding technologies were developed by taking advantage of DNA characteristics to extend the capability of multiplexing. Several methods such as DNA Exchange Imaging, Codetection by Indexing, Signal Amplification by Exchange Reaction, InSituPlex, and Digital Spatial Profiling (DSP) emerged. Among these methods, NanoString’s GeoMx® Digital Spatial Profiler (DSP) was among the first such platforms introduced into the market in 2019.
The GeoMx® Digital Spatial Profiler
GeoMx DSP is a multiplex, spatial multiomics platform for both proteins and mRNA that can be used on a variety of tissue samples such as fresh frozen or FFPE tissue sections. The GeoMx DSP technology is based on three basic premises:
- Oligonucleotide tags attached to antibody or in situ hybridization (ISH) probes act as barcodes for the readout of proteins or RNA expression
- Fluorescently-conjugated antibodies or ISH probes are used to stain and image samples
- UV light shined on a particular region of interest (ROI) on the tissue section releases the oligonucleotide tags so they can be collected and quantified. The abundance of these tags is then mapped back to the tissue location to allow spatial profiling of RNA and protein expression with the selected ROIs.
The GeoMx DSP platform can spatially profile tens to thousands of RNA molecules up to the whole transcriptome as well as 150+ proteins. The staining pattern of fluorescent labeled antibodies or ISH probes is used to help identify anatomical structures, tissue compartments, and cell types for profiling.
GeoMx DSP does not destroy the tissue sample and can be used to profile multiple ROIs. It is compatible with samples such as core needle biopsies, tissue microarrays, fresh frozen tissue, and FFPE tissue.
There are several advantages to using the GeoMx DSP for spatial profiling. The GeoMx DSP does not destroy the tissue sample and can be used repeatedly to profile multiple ROIs and is compatible with a variety of samples such as core needle biopsies, tissue microarrays, fresh frozen tissue, and the notoriously difficult to work with FFPE tissue. Another advantage of the GeoMx DSP platform is a flexible ROI selection strategy that can be customized for any number of tissue compartments and cell types. A key feature of the GeoMx DSP is an array of steerable reflective micromirrors called a digital micromirror device (DMD); these mirrors can be dynamically configured to profile an ROI of any shape or size on the tissue within the given field of view, including non-contiguous regions such as an entire population of one cell type. Consequently, the spatial resolution of the GeoMx DSP depends on the ROI selection strategy and sampling plan.
There are several GeoMx assays available off-the-shelf that include pre-validated antibody-based or RNA-based reagents that profile specific targets. Assays can even be customized to target exogenous markers, non-coding RNAs, and splice variants.
Applications of GeoMx DSP
Spatial multiomics with the GeoMx DSP has enabled discoveries in many research fields including oncology, immunology, neuroscience, and infectious disease. Some recent publications enabled by the GeoMx DSP platform are discussed below that highlight its translational capabilities.

Breast cancer is a highly heterogeneous disease across its histology, epidemiology, and molecular properties. Understanding the molecular diversity of breast cancer is therefore of utmost importance in determining the right therapy. As an example, the study done by McNamara et al. in 2021 used the GeoMx DSP platform to spatially resolve protein expression in the tumor and microenvironment compartments of archival FFPE tissue taken from a HER2+ neoadjuvant breast cancer biopsy. The study showed a change in expression levels of CD45, an immune biomarker, upon treatment that could predict patient outcomes.1McNamara KL, Caswell-Jin JL, Joshi R, Ma Z, Kotler E, Bean GR, Kriner M, Zhou Z, Hoang M, Beechem J, Zoeller J, Press MF, Slamon DJ, Hurvitz SA, Curtis C. Spatial proteomic characterization of HER2-positive breast tumors through neoadjuvant therapy predicts response. Nat Cancer. 2021 Apr;2(4):400-413. doi: 10.1038/s43018-021-00190-z. Epub 2021 Apr 8. PMID: 34966897; PMCID: PMC8713949.
Another study by Moutafi et al. investigated patients with advanced non-small-cell lung cancer (NSCLC) who failed to derive significant benefit from programmed cell death protein-1 (PD-1) axis blockade therapy, which is a standard treatment for multiple malignancies. Using the GeoMx DSP, the authors showed that CD44, a non-kinase transmembrane glycoprotein, was overexpressed in the tumor compartment, but not in the immune compartment, predicting clinical outcomes from PD-1 axis blockade.2Moutafi MK, Molero M, Martinez Morilla S, Baena J, Vathiotis IA, Gavrielatou N, Castro-Labrador L, de Garibay GR, Adradas V, Orive D, Valencia K, Calvo A, Montuenga LM, Ponce Aix S, Schalper KA, Herbst RS, Paz-Ares L, Rimm DL, Zugazagoitia J. Spatially resolved proteomic profiling identifies tumor cell CD44 as a biomarker associated with sensitivity to PD-1 axis blockade in advanced non-small-cell lung cancer. J Immunother Cancer. 2022 Aug;10(8):e004757. doi: 10.1136/jitc-2022-004757. PMID: 36002182; PMCID: PMC9413286.
In the field of neurobiology, the GeoMx DSP was utilized to understand the underlying neurodegenerative changes in resilient individuals who demonstrated neuropathologic changes consistent with Alzheimer’s disease, yet remained cognitively normal. Spatial expression analysis for proteins associated with CNS cell-typing or known neurodegenerative changes were done on hippocampal neurofibrillary tangle (NFT)-bearing neurons, non-NFT-bearing neurons, and their immediate neuronal microenvironments on FFPE tissue sections.3Walker JM, Kazempour Dehkordi S, Fracassi A, Vanschoiack A, Pavenko A, Taglialatela G, Woltjer R, Richardson TE, Zare H, Orr ME. Differential protein expression in the hippocampi of resilient individuals identified by digital spatial profiling. Acta Neuropathol Commun. 2022 Feb 14;10(1):23. doi: 10.1186/s40478-022-01324-9. PMID: 35164877; PMCID: PMC8842950. The study identified 11 proteins displaying differential expression in NFT-bearing neurons of the resilient individual compared to affected individuals, suggestive of an environment containing less energetic and oxidative stress, which in turn results in the maintenance of neurons and their synaptic connections.
Thus, these research publications demonstrate that GeoMx-based spatial strategies can be used for translational research, contributing to a more in depth understanding of biological mechanisms and disease.
References
- 1McNamara KL, Caswell-Jin JL, Joshi R, Ma Z, Kotler E, Bean GR, Kriner M, Zhou Z, Hoang M, Beechem J, Zoeller J, Press MF, Slamon DJ, Hurvitz SA, Curtis C. Spatial proteomic characterization of HER2-positive breast tumors through neoadjuvant therapy predicts response. Nat Cancer. 2021 Apr;2(4):400-413. doi: 10.1038/s43018-021-00190-z. Epub 2021 Apr 8. PMID: 34966897; PMCID: PMC8713949.
- 2Moutafi MK, Molero M, Martinez Morilla S, Baena J, Vathiotis IA, Gavrielatou N, Castro-Labrador L, de Garibay GR, Adradas V, Orive D, Valencia K, Calvo A, Montuenga LM, Ponce Aix S, Schalper KA, Herbst RS, Paz-Ares L, Rimm DL, Zugazagoitia J. Spatially resolved proteomic profiling identifies tumor cell CD44 as a biomarker associated with sensitivity to PD-1 axis blockade in advanced non-small-cell lung cancer. J Immunother Cancer. 2022 Aug;10(8):e004757. doi: 10.1136/jitc-2022-004757. PMID: 36002182; PMCID: PMC9413286.
- 3Walker JM, Kazempour Dehkordi S, Fracassi A, Vanschoiack A, Pavenko A, Taglialatela G, Woltjer R, Richardson TE, Zare H, Orr ME. Differential protein expression in the hippocampi of resilient individuals identified by digital spatial profiling. Acta Neuropathol Commun. 2022 Feb 14;10(1):23. doi: 10.1186/s40478-022-01324-9. PMID: 35164877; PMCID: PMC8842950.