A Spatial Twist on Biomarkers: Using Spatial Transcriptomics for Biomarker Discovery
Anyone who studies biology knows that living systems are highly complex, sophisticated entities, often containing redundant processes. Sorting out the mechanisms at play in any biological system is therefore aided by the availability of reliable guideposts for mapping out the details.
Objective points of reference, known as biomarkers, can serve as landmarks for evaluating progress toward a desired endpoint, making them key components of a successful research program.
Most people are familiar with traditional biomarkers such as hormone levels, weight, blood pressure, and body temperature. While effective at guiding physicians in diagnosis and treatment, they are not particularly precise, but are fine for initial diagnoses. The advent of precision medicine, not to mention molecular biology research, requires precise molecular biomarkers to act as objective guideposts. Modern biomarkers seek to deliver higher precision with gene expression signatures, or profiles, which are cell- or tissue-specific patterns of observed RNA transcripts.
Molecular biomarkers characterize specific cell types, as well as differentiate between various stages of diseases, bringing science closer to delivering on the promise of precision medicine.
With current technology, an entire transcriptome can be analyzed at all levels of an organism, from tissue down to subcellular compartments, providing exact information about which genes are expressed at a given time and place. The degree of information embedded in these gene expression profiles (GEPs) is immense and is derived from their complexity, making them highly robust and reliable. Fortunately, the power of bioinformatics helps navigate the massive amount of data generated when assessing modern biomarker changes within tissues under various conditions. Molecular biomarkers characterize specific cell types, as well as differentiate between various stages of diseases, bringing science closer to delivering on the promise of precision medicine. For this reason, biomarkers are a necessary part of molecular biology research.
 The CosMx SMI instrument.
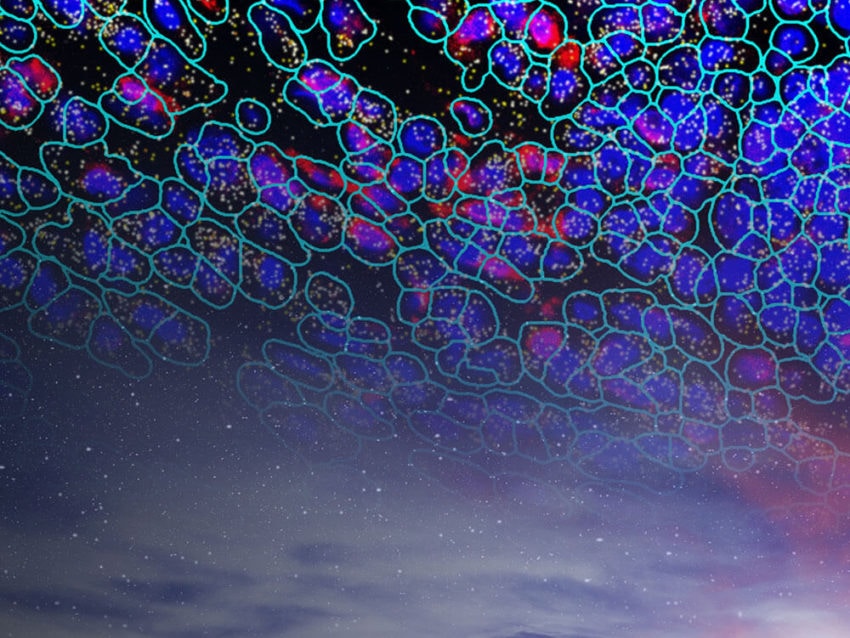
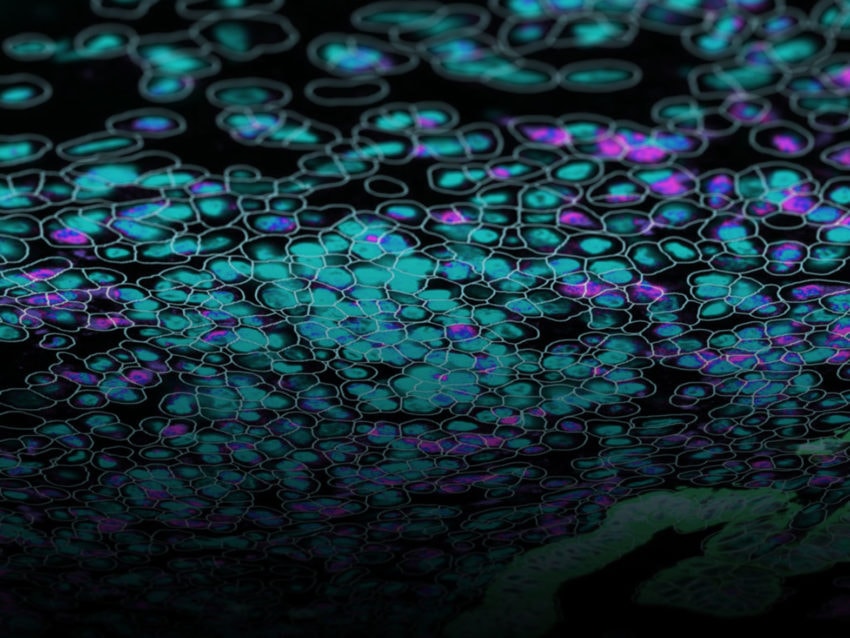
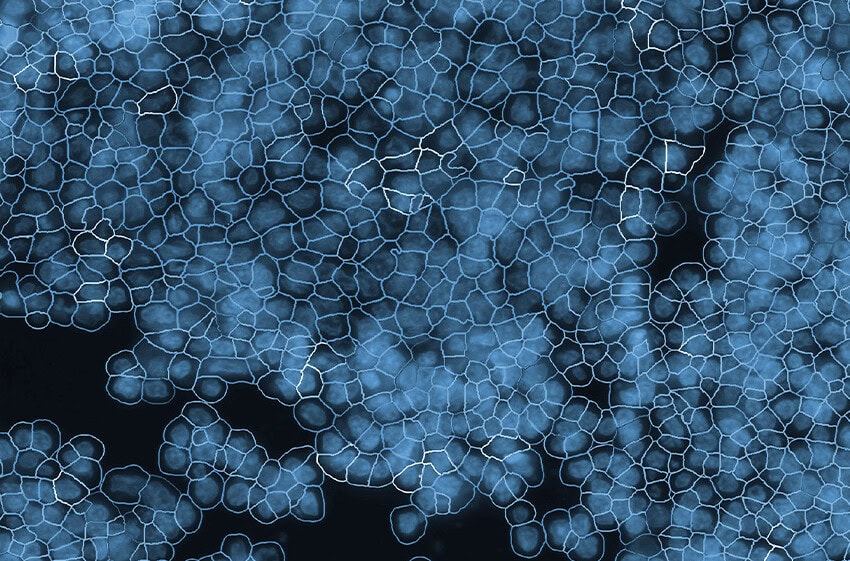
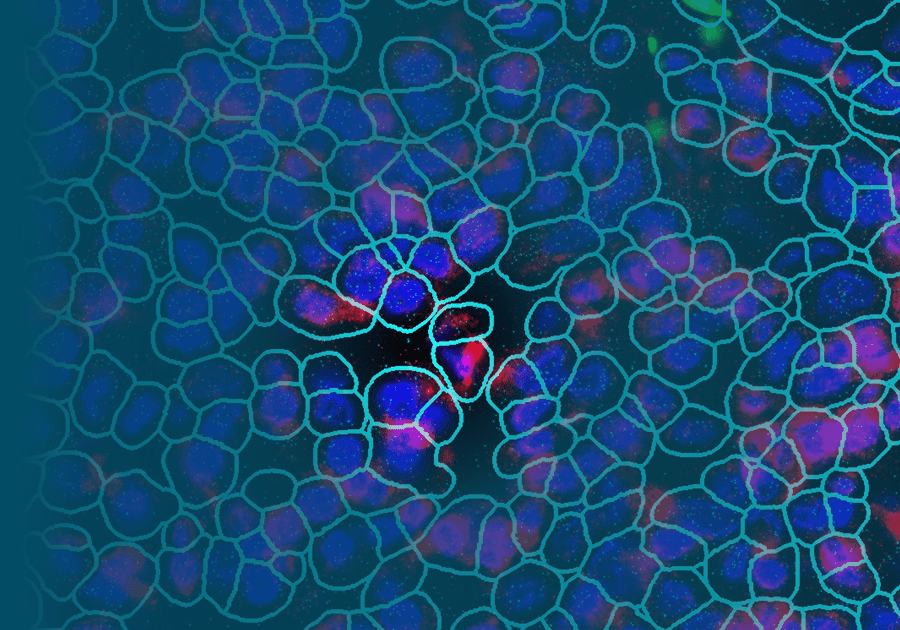
The CosMx SMI instrument.The advance in transcriptomics pushing biomarker discovery the farthest is the introduction of spatial context to GEP analysis. While earlier technologies, such as single cell RNA sequencing, identified genes for use as biomarkers for early-stage lung cancer, that information lacked context about the location those genes were expressed within the tissue architecture. Newer technologies now include spatial information in their analysis of GEPs. Additionally, some spatial biology platforms, such as NanoString’s CosMx® Spatial Molecular Imager (SMI), provide protein expression data along with transcriptomic information within that same spatial context. Dubbed spatial multiomics, CosMx SMI can identify new highly-specific biomarkers and also quantify changes in those biomarkers in response to treatments.
This article explores the biology of biomarkers, with a specific focus on the role of CosMx SMI. After discussing the basics of biomarkers, contributions of CosMx SMI to biology’s knowledge-base will be described, such as its use in characterizing cell types and identifying novel biomarkers.
What are biomarkers?
Biomarkers are a measurable property of a biological system indicating its current state of function. Some biomarkers are reference points for the overall state of the organism, such as blood pressure. Other biomarkers refine the focus to the state of one or more systems, such as the various available blood tests. Although good indicators of the overall state of a biological system, these biomarkers are too crude to precisely direct research or diagnosis. To achieve more precision, biomarkers are needed that examine cellular and subcellular levels. Reliable assays that can locate and quantify specific molecules are thus crucial to achieving that precision. For example, immunofluorescent assays that detect the presence of certain proteins help narrow the focus of subsequent research or diagnostic decisions. Similarly, detecting differences in transcriptional activity in specific cell types give valuable information for next steps. Molecular biomarkers are thus key for reliable and objective measures of a biological system’s current state.
A molecular biomarker that is proving highly valuable is the gene signature known as the Tumor Inflammation Signature (TIS). Composed of 18 different genes, TIS indicates the degree to which a patient’s T-cells have infiltrated their tumor and is used as a predictor of the patient’s response to anti-PD-1/PDL-1 immunotherapy.1Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, Piha-Paul SA, Yearley J, Seiwert TY, Ribas A, McClanahan TK. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017 Aug 1;127(8):2930-2940. doi: 10.1172/JCI91190. Epub 2017 Jun 26. PMID: 28650338; PMCID: PMC5531419. Evaluating the relative expression levels of the 18 genes represented in TIS imbues it with a degree of precision and reliability that sets it and other GEP biomarkers apart from systemic ones like blood pressure. It is this quantification and weighting of multiple genes that enables GEP biomarkers to predict a patient’s response to cancer treatment, such as pembrolizumab, with maximal accuracy.2Haddad RI, Seiwert TY, Chow LQM, Gupta S, Weiss J, Gluck I, Eder JP, Burtness B, Tahara M, Keam B, Kang H, Muro K, Albright A, Mogg R, Ayers M, Huang L, Lunceford J, Cristescu R, Cheng J, Mehra R. Influence of tumor mutational burden, inflammatory gene expression profile, and PD-L1 expression on response to pembrolizumab in head and neck squamous cell carcinoma. J Immunother Cancer. 2022 Feb;10(2):e003026. doi: 10.1136/jitc-2021-003026. PMID: 35217573; PMCID: PMC8883256.
How are biomarkers used to guide clinical decisions?
Molecular biomarkers give precise information about the state of a biological system. In clinics, therefore, they are becoming highly valued as an assessment of health status, enhancing and, in some cases, replacing systemic biomarkers such as blood pressure and weight. Indeed, their use in clinics is becoming increasingly common as associated costs have declined.
Biomarkers are being used at all levels of research to assess baseline and disease states of tissues. Indeed, certain biomarkers are now being assessed during clinical trials for approval in diagnosing clinical outcomes. Indeed, the TIS biomarker, mentioned above, was recently used in two Phase 1 clinical trials to aid in selecting immunotherapy-naïve cancer patients for treatment with either a combination anti-PD1 + ICOS agonist, or anti-PD1 alone. Not only did TIS accurately predict patient response to treatment, but it was also instrumental in the discovery of a novel biomarker that correlated with both high TIS scores and good treatment-response rates.3Yap, et.al. ASCO-SITC 2020. DOI: 10.1200/JCO.2020.38.5_suppl.14 Journal of Clinical Oncology 38, no. 5_suppl (February 10, 2020) 14-14.
Biomarkers are also used as potential risk factors for developing certain types of cancer. For example, probably the most well-known biomarkers are the BRCA genes associated with risk of breast cancer. Certain mutations in the BRCA genes disrupt the function of the associated DNA repair protein product, leading to increased DNA damage, a major step toward tumorigenesis. Biomarkers such as these BRCA mutations thus can guide patients to pursue additional screening to determine their risk of further disease development.
Using the SERT gene mutation as a biomarker not only helps physicians choose the appropriate treatment, but can also help avoid negative outcomes.
Biomarkers are also used by psychiatrists to predict patient response to pharmacological treatment. For example, patients with a certain mutation in the serotonin transporter (SERT) gene do not respond to frontline selective serotonin reuptake inhibitors (SSRIs), and may also experience severe side effects from these medications. Using this SERT mutation as a biomarker thus not only helps physicians choose the appropriate treatment, but can also help avoid negative outcomes.
What is spatial transcriptomics and how does it relate to biomarker discovery?
For biomarkers to facilitate better individual medical care, relevant biomarkers must first be characterized. Complex molecular biomarkers, such as gene expression profiles, require sophisticated technologies to gather and analyze the large amounts of data involved.
Preserving the cellular location of expressed RNAs within a tissue helps explain their biological role.
One technology accelerating the identification and use of biomarkers clinically is spatial transcriptomics (sometimes called spatial multiomics). Spatial transcriptomics surpasses previous next-generation sequencing techniques, such as single cell RNA seq, by framing tissue and cellular transcriptomic information in a spatial context, providing more precise analysis of the cellular biology and interactions involved in a given system. Characterization of gene expression spatial context is made possible by barcoded hybridization probes that identify transcripts while keeping the target tissue intact. Preserving the cellular location of expressed RNAs within a tissue helps explain their biological role. Additionally, spatial transcriptomics can be used to analyze protein expression for simultaneous characterization with RNA expression. The hybridization protocol used in spatial transcriptomics for screening barcoded probes removes cDNA library preparation or PCR amplification steps needed in single cell RNA seq that introduce potential errors.
As an example, spatial transcriptomics gives oncologists the ability to identify novel, cell-specific biomarkers in tumor cells. For instance, the epidermal growth factor receptor (EGFR) gene containing certain mutations was identified as a tumor cell-specific biomarker, the presence of which predicts a patient will respond well to EGFR inhibitor treatment . Using spatial transcriptomics to identify the EGFR biomarker in thus informs oncologists about their treatment options, and highlights the role this technology in precision medicine.
How does CosMx SMI work to provide information on biomarkers?
NanoString has a complete spatial transcriptomics solution that includes CosMx SMI and an associated cloud based bioinformatics package, AtoMx® Spatial Informatics Platform (SIP). Using the CosMx SMI, users can use formalin-fixed paraffin embedded or fresh frozen samples to detect up to 1000 RNA and 64 protein samples at once. The multiomic output allows users to easily visualize cell boundaries and identify regions of interest (ROI). AtoMx SIP also will work with NanoString’s other spatial biology instrument, the GeoMx® Digital Spatial Profiler (DSP).
Used in combination, GeoMx DSP and CosMx SMI are valuable tools for biomarker discovery. GeoMx DSP provides an overview of the spatial multiomics in a tissue, guiding research to ROI for further investigation. More refined analysis of specific cells and cellular compartments can then be performed with with CosMx SMI, examining biomarker expression patterns to elucidate information about the functional organization of tissue microenvironments and discover novel cell specific biomarkers.
How has CosMx SMI been used to identify biomarkers?
Since spatial transcriptomics characterizes gene expression networks within cells and tissues, researchers can generate cell atlases based on those expression patterns. Furthermore, atlases for various conditions can be generated, such as separate ones for healthy versus cancerous tissues.
One group using both the GeoMx DSP and the CosMx SMI examined differences in biomarker expression among three different types of skin cancer (basal cell carcinoma, squamous cell carcinoma, and melanoma) to generate atlases of each. The atlases were then used to characterize the involvement of PD-1+ immune cells and PD-L1+ cancer cells in cell-cell interactions, especially at the tumor boundary. In this way, spatial transcriptomics expands understanding of these interactions, decoding the causes of cancer and leading to more effective treatments.4Grice LF, Jin X, Tran M, Mulay O, Killingbeck E, Gregory M, Teoh SM, Devitt K, Kulasinghe A, Leon M, Murphy S, Warren S, Khosrotehrani K, Stark M, Frazer I, Kim Y, Nguyen Q. Building a spatial single-cell multi-omics atlas and cellular interactome for skin cancer. 2021 5Kiuru M, Kriner MA, Wong S, Zhu G, Terrell JR, Li Q, Hoang M, Beechem J, McPherson JD. High-Plex Spatial RNA Profiling Reveals Cell Type‒Specific Biomarker Expression during Melanoma Development. J Invest Dermatol. 2022 May;142(5):1401-1412.e20. doi: 10.1016/j.jid.2021.06.041. Epub 2021 Oct 23. PMID: 34699906; PMCID: PMC9714472.
Spatial transcriptomics instruments like CosMx SMI can improve understanding of biomarkers in tissues and the response of those tissues to various treatments.
Additionally, spatial transcriptomics instruments such as CosMx SMI can be used to improve understanding of biomarkers in healthy tissues and the response of those tissues to various treatments. For example, spatial transcriptomics can help answer questions such as: do different cells in the same tumor respond differently to treatment?; are some cells more resistant to treatment than others?; do benign cells behave differently in the tumor microenvironment? Answering such questions will lead to better treatment therapies and enhance our understanding of the variations in treatment response of patients with the same type of cancers. In this way, spatial transcriptomics will accelerate cancer and disease treatments in the very near future.
Biomarkers as molecular guideposts
Medical professionals have long used biomarkers in their assessments of their patients’ health status. Adopting molecular biomarkers as the standard of care allows physicians to more precisely diagnose and treat their patients. The use of spatial transcriptomics and similar technologies will continue to expand the available repertoire of biomarkers, advancing knowledge of biological systems, both healthy and diseased. NanoString’s suite of spatial biology instruments is leading the way toward that future.
The CosMx® SMI and decoder probes are not offered and/or delivered to the Federal Republic of Germany for use in the Federal Republic of Germany for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof in a method used in fluorescence in situ hybridization for detecting a plurality of analytes in a sample without the consent of the President and Fellows of Harvard College (Harvard Corporation) as owner of the German part of EP 2 794 928 B1. The use for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof is prohibited without the consent of the President and Fellows of Harvard College (Harvard Corporation).
References
- 1Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, Piha-Paul SA, Yearley J, Seiwert TY, Ribas A, McClanahan TK. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017 Aug 1;127(8):2930-2940. doi: 10.1172/JCI91190. Epub 2017 Jun 26. PMID: 28650338; PMCID: PMC5531419.
- 2Haddad RI, Seiwert TY, Chow LQM, Gupta S, Weiss J, Gluck I, Eder JP, Burtness B, Tahara M, Keam B, Kang H, Muro K, Albright A, Mogg R, Ayers M, Huang L, Lunceford J, Cristescu R, Cheng J, Mehra R. Influence of tumor mutational burden, inflammatory gene expression profile, and PD-L1 expression on response to pembrolizumab in head and neck squamous cell carcinoma. J Immunother Cancer. 2022 Feb;10(2):e003026. doi: 10.1136/jitc-2021-003026. PMID: 35217573; PMCID: PMC8883256.
- 3Yap, et.al. ASCO-SITC 2020. DOI: 10.1200/JCO.2020.38.5_suppl.14 Journal of Clinical Oncology 38, no. 5_suppl (February 10, 2020) 14-14.
- 4Grice LF, Jin X, Tran M, Mulay O, Killingbeck E, Gregory M, Teoh SM, Devitt K, Kulasinghe A, Leon M, Murphy S, Warren S, Khosrotehrani K, Stark M, Frazer I, Kim Y, Nguyen Q. Building a spatial single-cell multi-omics atlas and cellular interactome for skin cancer. 2021
- 5Kiuru M, Kriner MA, Wong S, Zhu G, Terrell JR, Li Q, Hoang M, Beechem J, McPherson JD. High-Plex Spatial RNA Profiling Reveals Cell Type‒Specific Biomarker Expression during Melanoma Development. J Invest Dermatol. 2022 May;142(5):1401-1412.e20. doi: 10.1016/j.jid.2021.06.041. Epub 2021 Oct 23. PMID: 34699906; PMCID: PMC9714472.