What is transcriptomics?
The transcriptome is the sum of all the RNA in an organism’s cells and tissues being actively transcribed at any given time from genomic DNA. It includes protein-coding messenger RNA (mRNA) and a variety of noncoding RNA such as circular RNA (CircRNA), microRNA (miRNA), and long-noncoding RNA (lnRNA). Transcriptomics, therefore, is the study of the complete set of RNA transcripts that are produced from genomic DNA in cells or tissues under specific circumstances.
Transcription profiling follows the changes in gene expression that occur over time, in different states of health and disease, or in response to some kind of stimuli such as drug treatment. Today transcriptomic analysis is an integral part of most genomic studies of biological processes and disease as it provides a link between genotype and phenotype.
Over the past decade, the study of transcriptomics has paved the way for spatial biology (also known as spatial genomics or spatial transcriptomics). Biology is inherently spatial: understanding how genes are expressed and interconnected in three-dimensional space is critical to gain insights into the proper functioning of many biological processes. Thus, spatial transcriptomics — the study of the transcriptome with spatial context — is allowing researchers to measure and visualize gene expression with a spatial context within individual cells or tissues.
Spatial transcriptomics
Spatial transcriptomics looks at the transcriptomic fingerprint or net spatial gene expression within intact tissues during development and disease. Today, spatial transcriptomics technologies primarily use high-throughput next-generation sequencing (NGS) methods to analyze the expression of multiple transcripts in different physiological or pathological conditions, enabling researchers to explore the molecular mechanisms underlying phenotypes, distinguish healthy and diseased states, and elucidate the etiology of disease.
Spatial proteomics
Predicting protein expression and spatial location based solely on mRNA levels has proven to be unreliable as many biological pathways are regulated at the level of post-translational modification and localization. Knowledge of the spatial localization of proteins within tissues is critical to understanding how protein function and trafficking between cells is regulated. Towards this aim, spatial proteomics methods capture the localization of proteins and their dynamics at the tissue or single cell level, contributing to a more complete understanding of molecular biology.
The GeoMx® Digital Spatial Profiler: A multiomics platform
Spatial multiomics integrates the methodologies used to study the whole genome (genomics), RNA expression (transcriptomics), protein abundance (proteomics), metabolites (metabolomics), and the epigenome (epigenomics) with spatial context to provide a comprehensive understanding of various complex cellular and molecular events within intact tissues.
NanoString’s GeoMx DSP is a spatial multiomic platform that profiles both RNA and protein expression in two dimensions in a wide variety of tissue sections, including formalin fixed, paraffin embedded (FFPE) or fresh frozen tissue. This approach relies on the quantification of RNA or proteins using oligonucleotide tags that are attached to affinity reagents such as antibodies or RNA probes through a photocleavable linker.
The affinity reagents are then hybridized onto a slide-mounted tissue section that is also stained with fluorescent antibodies or in situ hybridization (ISH) probes to visualize the tissue. The tissue is then imaged using fluorescence microscopy and UV light is projected onto selected regions of interest (ROI) to release the oligonucleotide tags from each ROI. These tags are collected and subsequently counted digitally using the nCounter® Analysis System or NGS.

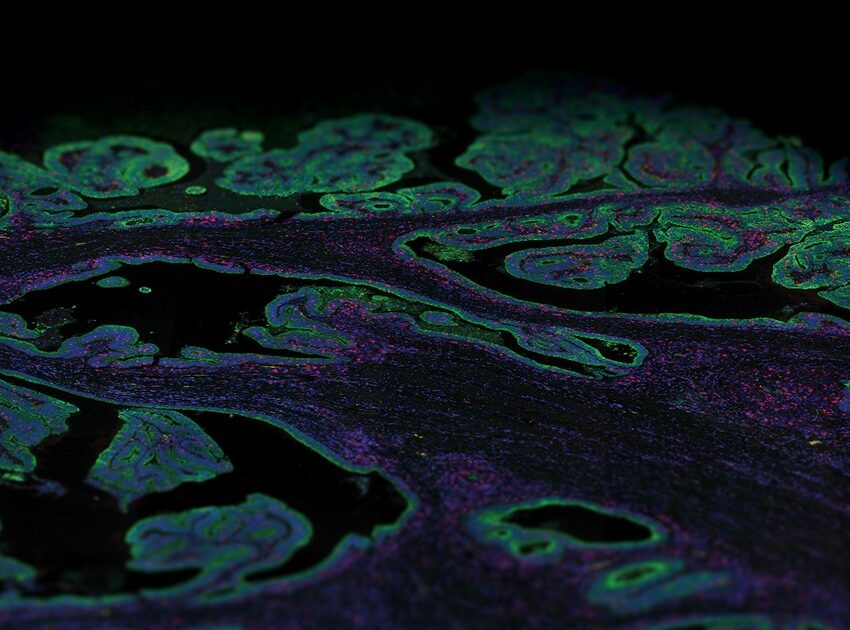
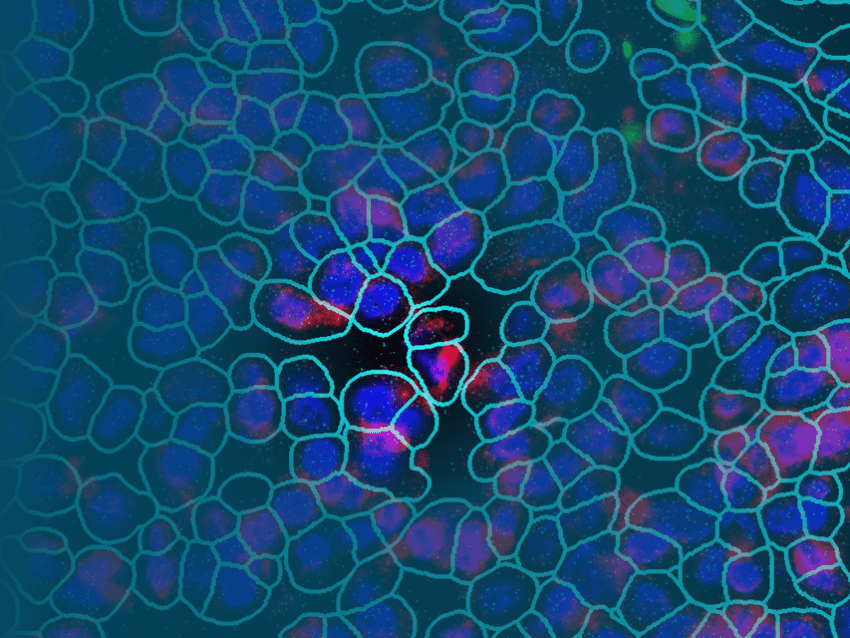
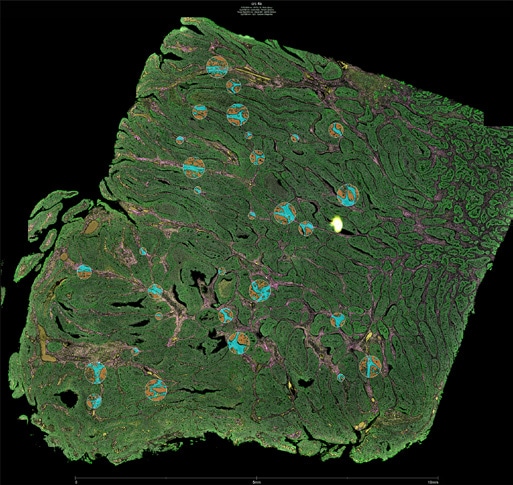
GeoMx allows for flexible and customizable ROI selection of different shapes or sizes, including shapes that trace out the boundaries of different histological features or specific cell types.
The GeoMx DSP instrument uses a digital micromirror device to select ROIs down to 10 microns in diameter; the DMD contains an array of tiny mirrors that can be oriented independently to shine UV light in a specific pattern on the tissue. This allows for flexible and customizable ROI selection of different shapes or sizes, including shapes that trace out the boundaries of different histological features or specific cell types.
GeoMx DSP can also profile a given cell type population within the field of view, even if the cells are discontinuous: the DMD can shine UV light on each cell simultaneously to collectively capture the oligonucleotide probes bound to each selected cell within the tissue section. Off-the-shelf RNA assays are available that profile upwards of 20,000 transcripts, essentially the whole human or mouse transcriptome or targeted groups of 96 RNAs. Protein assays include probes for 150+ targets, and you can even spatially profile RNA and protein simultaneously from the same tissue section!
Applications of the GeoMx DSP
The GeoMx DSP spatial multiomics platform can be applied to investigate a number of biological processes in the field of oncology, neuroscience, immunology, and infectious disease. FFPE tissue is the routine form of preservation in clinical practice for biopsies, making the GeoMx DSP the ideal spatial biology platform for translational and clinical research.
GeoMx DSP’s support for FFPE tissue makes it the ideal spatial biology platform for translational and clinical research.
Furthermore, the flexible ROI selection strategy that uses tissue staining of morphological markers as a guide is very advantageous for identifying different tissue compartments representing anatomical structures in organs or disease pathology. Recent studies have shown the feasibility of using the GeoMx DSP platform in studying different types of tumors, characterizing tissue heterogeneity and the immune infiltrate, and identifying/validating biomarkers.
The GeoMx DSP provides a useful toolbox for tissue phenotyping or constructing spatial tissue atlases that catalog the biology of all kinds of species, organisms, and infectious diseases. For example, GeoMx DSP was used to create an immune response atlas for COVID-19 during the 2020 pandemic.