A Strategic Attack Against Neurological Diseases
Neurological diseases are often multifactorial, brought on by a genetic and environmental tug-of-war and resulting in multifaceted clinical presentations. Such etiological complexity has hindered treatment progress and has necessitated well-thought-out and systematic strategies of attack to mitigate their global burden.
When designing such strategies, one needs to consider the following:
DATA TYPE — What to measure
With a plethora of biomedical “big data”, researchers are faced with the challenge of selecting molecular data types that can be readily probed for novel and clinically informative insights. The transcriptome, which encompasses the entire set of assorted ribonucleic acid (RNA) molecules, is extensively interrogated in such efforts. It conveys a steady-state snapshot of total gene activity and, with the advent of high throughput profiling technologies, can reveal spatiotemporally-resolved disease patterns.
DATA SOURCE — Where to measure from
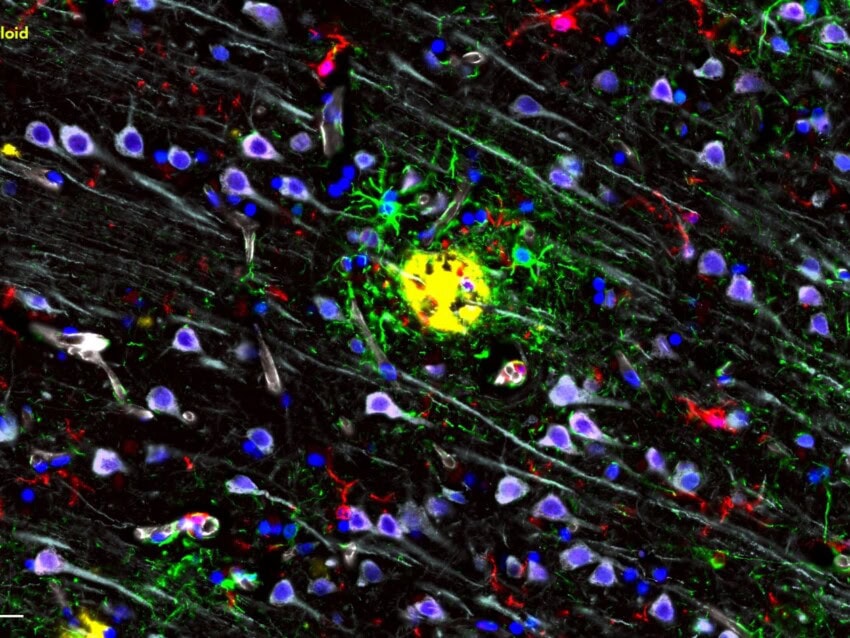
We now need to identify an adequate biological source for the data type in mind. Postmortem human brain tissue represents a direct medium for examining divergent transcriptional programming in neurological diseases but can be difficult to access. Animals are therefore used as the gold standard for in vivo experimental modeling of human diseases. Transgenic models are developed to closely reflect disease biology, exhibiting key pathophysiological features that allow thorough testing of mechanistic hypotheses and therapeutic efficacy. Reconstructing complex disease states is, nonetheless, difficult, and animal-to-human results are often lost in translation. New models are in development to more accurately recapitulate human disease. In this webinar, Dr. Gregory Carter shows how human disease-relevant gene expression assays can be leveraged to screen for murine strains that recapitulate late-onset Alzheimer’s disease phenotypes.
DATA ANALYSIS — How to measure
A meticulous selection of analytical technique is critical to the effective extraction of information from collected data. Pathogenic deregulations at the transcriptome level are commonly inferred by observing genome-wide changes in messenger RNA (mRNA) levels. Significant strides have been made in identifying gene expression correlates of neurological diseases. Nonetheless, the poor reproducibility of such findings, due to sample-, platform-, and analysis-associated variability, continues to pose a translational barrier. Furthermore, lowly-expressed genes that may be essential to disease signature are hard to capture when unbiasedly measuring tens of thousands of genes from limited biological material. Such limitation can be alleviated through targeted expression profiling of carefully curated genes using a wide range of sample types. In this webinar, Dr. Kelly Miller discusses how the NanoString® nCounter® platform, along with the nCounter Neuropathology and Neuroinflammation Panels, can improve the detection of genes with differential expression patterns in neuropathology.
The etiology of complex diseases cannot be pinpointed to a single origin; rather, it manifests as global and patient-specific perturbations to a complex system of cells and genes. By applying the “what, where, and how to measure” strategy, researchers can better track down disease origins, and a fortiori, those involving multiple components and biological layers, as seen in neurological diseases.