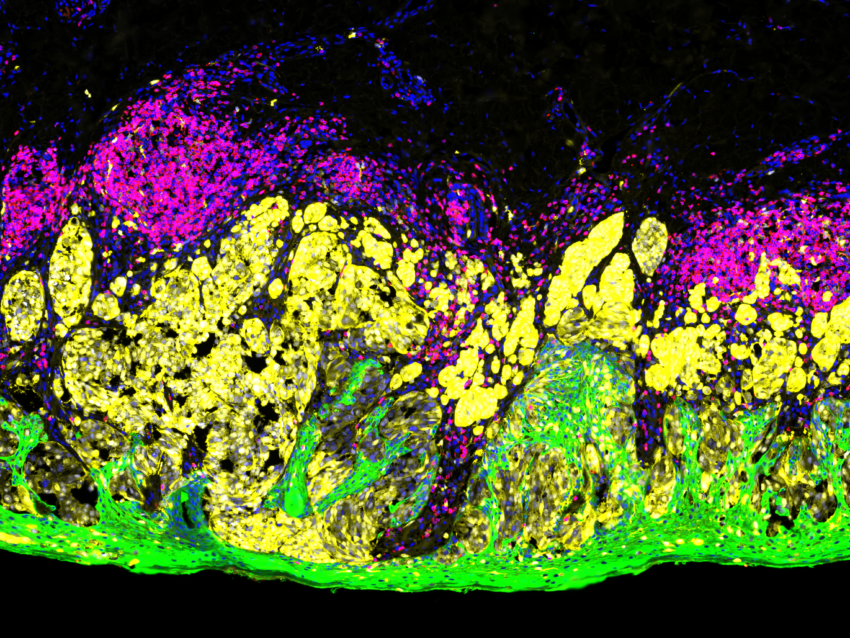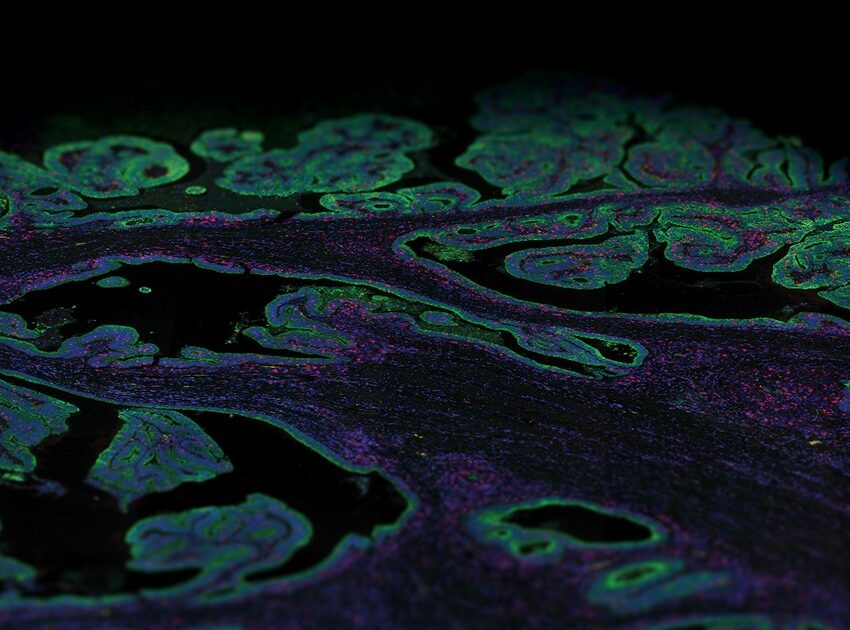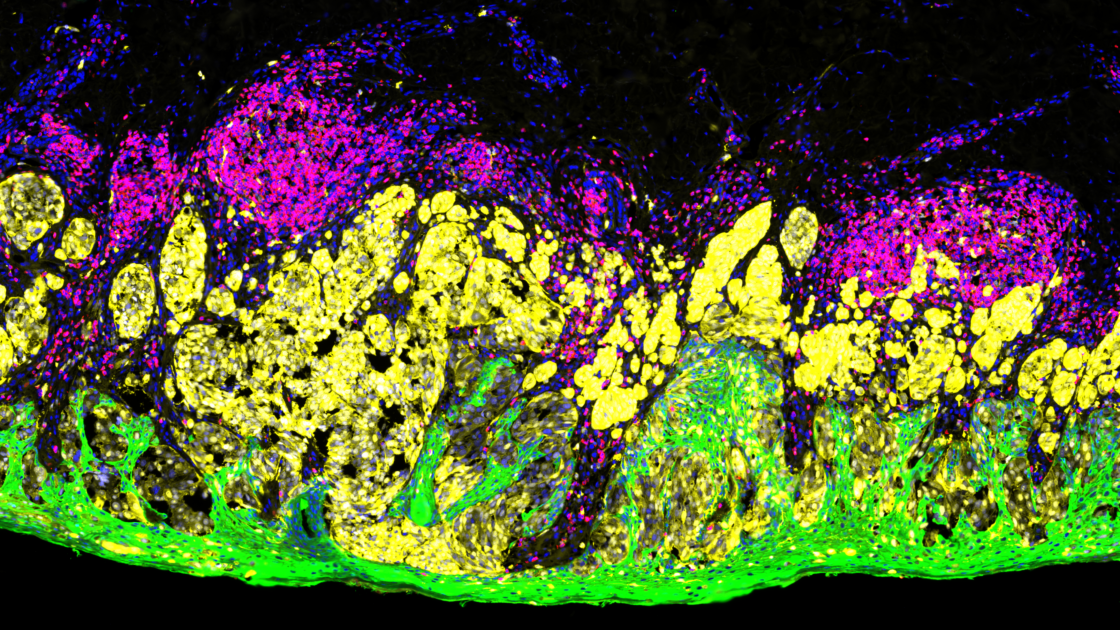
How is immunofluorescence staining done?
Immunofluorescence (IF) staining is a widely used technique that allows for the detection and localization of antigens in any given tissue or cell type.
IF methods use combinations of specific antibodies or in situ hybridization (ISH) probes tagged with fluorophores that are visualized with microscopy, and can be performed on a variety of sample types ranging from cells in culture or suspension to tissue samples.
Over the last decade, IF staining has been further developed to work on formalin-fixed, paraffin-embedded (FFPE) tissue sections, making available a wealth of valuable clinical material that can now be used routinely for molecular analysis. Additionally, the development of RNA and protein profiling technologies, many of which are based on IF staining, is advancing our knowledge of the spatial relationship between different target proteins in the context of tissue architecture.
Immunofluorescence staining on FFPE tissue
To perform IF staining, it is crucial that the biological specimen is well-preserved, and that the target antigens are made accessible to antibodies. The workflow for IF staining involves fixing the tissue followed by antigen retrieval and blocking. Finally, the sample is incubated with appropriate fluorescently tagged antibodies or ISH probes and visualized for fluorescence with various microscopy techniques
Fixation of tissue
Fixation is an essential preliminary step in IF staining to immobilize target antigens without disturbing cellular architecture, allowing maximum access to any targeted cellular components. FFPE tissue samples are fixed using formaldehyde, a commonly used fixative that acts by chemically cross-linking free amine groups on proteins and can partially permeabilize membranes. After fixation, the tissue is embedded in a paraffin wax block. Tissue slices are then cut off from the paraffin block and mounted on a microscopic slide. The next step involves deparaffinization and rehydration so that only the tissue section remains on the slide for subsequent antigen retrieval and IF staining. Xylene is commonly used for deparaffinization, followed by washes of ethanol and distilled water for rehydration.
Antigen retrieval and blocking
Fixation can modify the antigen targets due to the cross-linking of free amines that can mask the target epitope. Therefore, an antigen retrieval step that cleaves the cross-links formed during fixation is necessary to restore the reactivity of the target antigens. Retrieval agents can use either enzymes such as proteases or the application of heat and pressure to restore antigenicity. After this step, the samples are incubated with a blocking agent to suppress the background from direct interaction of the fluorescent dyes with the samples, as well as non-specific binding. Blocking reagents should not have an affinity for the target epitope and have a high binding rate to non-target reactive sites.
Antibodies
Following antigen retrieval and blocking, IF staining with antibodies can be performed with either of two methods: direct or indirect detection. Direct IF detection utilizes fluorescently conjugated monoclonal antibodies and only one antibody molecule that is conjugated to 4-6 fluorescent dyes may bind to its target antigen. Indirect IF detection is a two-step process: first, an unlabeled primary antibody is bound to the target antigen followed by the binding of multiple fluorescently conjugated secondary antibodies to the primary antibody.
The indirect method results in signal amplification and higher sensitivity compared to direct detection as each secondary antibody is conjugated to 4-6 fluorescent dyes, thereby increasing the number of fluorophores associated with the target antigen. However, direct IF offers the advantage of being able to stain a sample with multiple primary antibodies from the same host species simultaneously. Direct and indirect methods can be combined to exploit the advantages of both methods.
Development of multiplex immunofluorescence staining
One of the main disadvantages of traditional IF staining is that it only allows the detection of a single biomarker per tissue section. Over the last decade, several multiplex IF staining technologies have evolved that detect multiple biomarkers from one tissue section. These multiplex IF methods still rely on direct or indirect detection of antigens utilizing a fluorescence microscope but are now fully automated using diverse detection methodologies.
NanoString’s GeoMx® Digital Spatial Profiler (DSP) is an example of such a multiplex platform that characterizes spatial expression of 100s to 1000s of RNA and/or protein targets from different sample types, including FFPE tissue sections. The GeoMx DSP platform uses oligonucleotide tags that are attached to antibodies or RNA probes through a photocleavable linker. These labeled probes then bind to their targets on a slide-mounted tissue sample that is also stained with immunofluorescent antibodies or ISH probes to visualize the sample and identify tissue structure. These tissue structures identified with morphology markers help in selecting different regions of interest (ROIs) on the tissue to be analyzed. UV light is then shined sequentially on each ROI selected, triggering the release of the oligonucleotide tags. These tags are collected and counted digitally using either the nCounter® Analysis System or high-throughput next-generation sequencing (NGS). Expression profiles for each ROI are then digitally mapped back to the tissue image and ROIs to get a spatial map of RNA and/or protein expression.