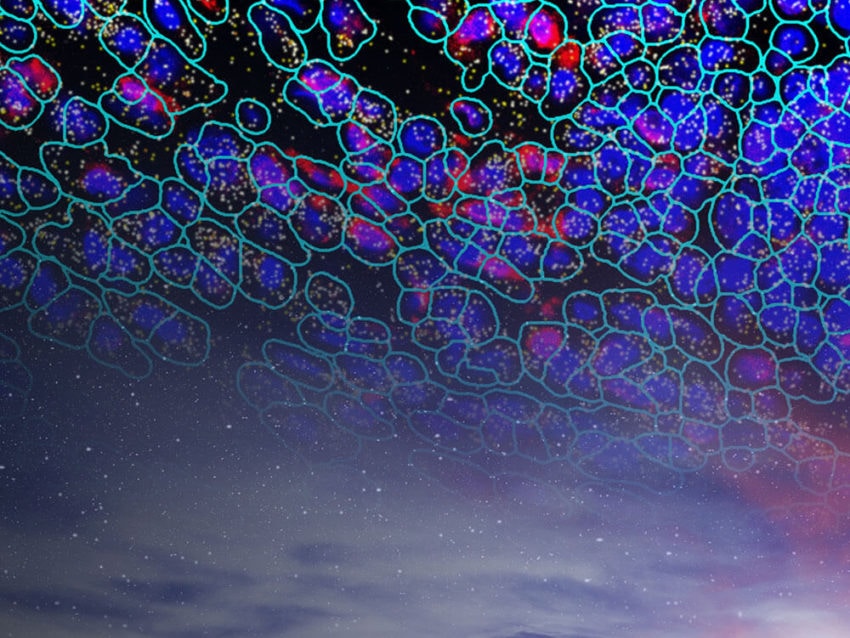
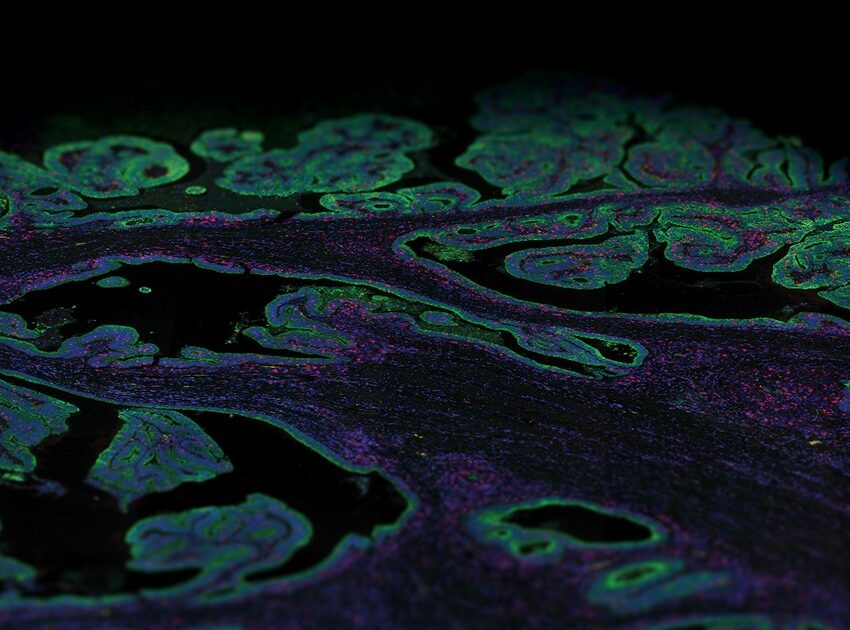
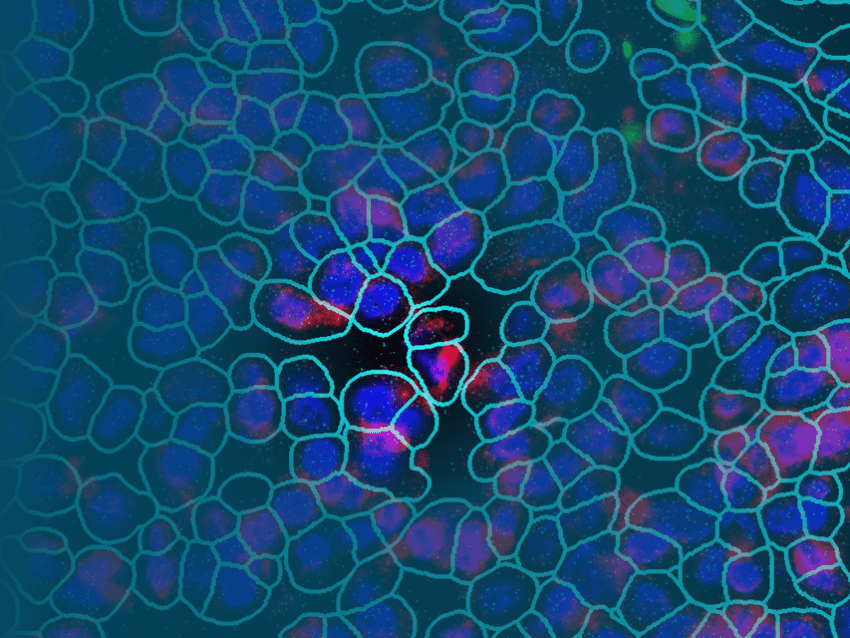
Single-Cell Spatial Surpasses Single-Cell Sequencing
Modern biology research is deeply interested in discovering the fundamental mechanics that operate within an organism of interest. Since the fundamental unit of all living organisms is the cell, it makes sense that researchers place a major focus on the inner workings of cells. The molecules responsible for cellular functions are therefore an intense area of study in nearly every field of biology today.
The key molecule of cellular function is DNA. Containing the instructions for all the cellular functions of an organism, DNA is analogous to a cellular recipe book. The “recipes” contained within the DNA encode RNAs, which in turn direct protein production. RNA and protein, in turn, comprise nearly all the structures and functions in a cell, meaning that proper cellular and organismal function rely on the appropriate production of these molecules. Production of RNA and protein is collectively known as gene expression. Analysis of gene expression plays a critical role in understanding the biological function of any cell, tissue, or organism.
Gene expression analysis most often refers to characterizations of RNA transcript levels, since RNA transcription always precedes protein production. Methods of RNA analysis consist of hybridization, amplification, or sequencing techniques, all of which are well-proven. However, for a long time, only hybridization techniques could give information about RNA levels and localized expression, although not at the same time. The introduction of single-cell sequencing — a combination of PCR, sequencing, and microfluidics — made it possible to obtain DNA or RNA sequence information from isolated single cells. Adding an initial reverse transcription step to the PCR part (rt-PCR) of single-cell sequencing enabled analysis of single-cell transcriptomes in a technique known as single-cell RNA sequencing (scRNA seq). This article discusses single-cell sequencing, particularly single-cell RNA seq, and the ways scRNA seq can advance research. It also discusses how the spatial biology platforms GeoMx® Digital Spatial Profiler and CosMx® Spatial Molecular Imager can provide information above and beyond that gleaned from scRNA seq.
What is single-cell sequencing?
Single-cell sequencing is a method in which cells are dissociated from a tissue and isolated to obtain sequence information of their individual DNA or RNA. Previously, sequence information for a tissue required tissue homogenization, effectively pooling the DNA or RNA from all cell types in the tissue. However, individual cell types in a tissue express different RNAs, and, depending on the tissue, may also have DNA differences. Gaining insight into cellular functions and cell-cell interactions in a tissue requires information about the responsible cell types. Single-cell sequencing provides that information.
For example, single-cell sequencing can identify any DNA mutations present in individual tumor cells, thus guiding studies of which genomic mutations may cause malignancy.1Li Y, Jiang X, Zhong M, Yu B, Yuan H. Whole Genome Sequencing of Single-Circulating Tumor Cell Ameliorates Unraveling Breast Cancer Heterogeneity. Breast Cancer (Dove Med Press). 2022 Dec 28;14:505-513. doi: 10.2147/BCTT.S388653. PMID: 36597488; PMCID: PMC9805725. scRNA seq can generate RNA profiles of specific cell types, also obtaining quantitation data for the transcripts identified.2Chan TW, Fu T, Bahn JH, Jun HI, Lee JH, Quinones-Valdez G, Cheng C, Xiao X. RNA editing in cancer impacts mRNA abundance in immune response pathways. Genome Biol. 2020 Oct 26;21(1):268. doi: 10.1186/s13059-020-02171-4. PMID: 33106178; PMCID: PMC7586670. Single-cell sequencing of DNA and RNA is highly informative for understanding the functions of various cell types. However, the initial tissue dissociation step required for single-cell sequencing disrupts any three-dimensional spatial context, thus lacking any information about cell-cell interactions. Regardless, single-cell sequencing can still provide valuable data for guiding further research.
Why is single-cell sequencing useful?
Single-cell sequencing is valuable technology due to the cellular heterogeneity of tissues. Human skin, for instance, consists of multiple layers, each characterized by different cell types necessary for proper skin function. Driving each cell type’s specific properties is differential gene expression.
Assessing a single cell’s RNA expression profile can identify genes for further study. For instance, a pioneering study in the field of organoid development used scRNA seq to assess the distribution of cell types found in mouse intestine organoids, leading to the identification of a novel biomarker, Reg4, for enteroendocrine cells, a rare population of hormone-producing intestinal cells.3Grun D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525(7568):251–5. The Reg4 biomarker discovered by scRNA seq can now be used as a tool to identify enteroendocrine cells in a heterogeneous population, helping gain a fuller understanding of those cells’ biological function. Additionally, Reg4 can be used to localize enteroendocrine cells in a spatial context. In this way, single-cell sequencing helps guide research decisions by identifying relevant DNA or RNA players.
Why does tissue context matter for gene expression?
Determining the tissue context for gene expression profiles matters because individual cells do not function alone in multicellular organisms. In fact, proper function of a multicellular organism requires active communication between cells in a tissue. Known as cell-cell interactions, these communications often depend on the presence of the appropriate gene products. Conversely, malfunction of biological systems, such as in tumor formation, can also rely on cell-cell interactions.4Kumar
Understanding where and when cells express specific genes elucidates the mechanisms of biological function.
In a study of ligand-receptor interactions in six types of tumors from syngeneic mice, scRNA seq data was used to characterize cell-cell communications across all cells in the tumor microenvironment based on their gene expression profiles. Study results showed this approach accurately assessed cell-cell communication across mouse tumor models.4Kumar MP, Du J, Lagoudas G, Jiao Y, Sawyer A, Drummond DC, Lauffenburger DA, Raue A. Analysis of Single-Cell RNA-Seq Identifies Cell-Cell Communication Associated with Tumor Characteristics. Cell Rep. 2018 Nov 6;25(6):1458-1468.e4. doi: 10.1016/j.celrep.2018.10.047. PMID: 30404002; PMCID: PMC7009724. A similar approach examined ligand-receptor interactions in human metastatic melanoma tumor microenvironment and showed similar results.4Kumar MP, Du J, Lagoudas G, Jiao Y, Sawyer A, Drummond DC, Lauffenburger DA, Raue A. Analysis of Single-Cell RNA-Seq Identifies Cell-Cell Communication Associated with Tumor Characteristics. Cell Rep. 2018 Nov 6;25(6):1458-1468.e4. doi: 10.1016/j.celrep.2018.10.047. PMID: 30404002; PMCID: PMC7009724. However, the absence of spatial information in this study prevented the determination of the spatial relationship of the cell types required for the membrane-bound receptor-ligand interactions identified. The authors suggest use of multiplexed fluorescently-labeled antibodies to validate the spatial co-localization of membrane-bound components.4Kumar MP, Du J, Lagoudas G, Jiao Y, Sawyer A, Drummond DC, Lauffenburger DA, Raue A. Analysis of Single-Cell RNA-Seq Identifies Cell-Cell Communication Associated with Tumor Characteristics. Cell Rep. 2018 Nov 6;25(6):1458-1468.e4. doi: 10.1016/j.celrep.2018.10.047. PMID: 30404002; PMCID: PMC7009724.
Inclusion of spatial localization in the analysis of a cell’s gene expression profile therefore uncovers the functionality of the relevant genes, facilitating a better understanding of the biological processes. In other words, understanding where and when cells express specific genes elucidates the mechanisms of biological function.
The value of single-cell sequencing in a spatial context
The field of organoid development is a fantastic example of single-cell sequencing use. Organoids are complex 3D cell cultures designed to mimic the properties of the analogous organs, including the presence of the appropriate cell types. When culturing organoids, technology such as scRNA seq is used to compare organoid models with the model organ. To date, organoid liver, intestine, endometrium, epididymis, biliary epithelium, salivary gland, and heart tissues have been cultured.5Smirnov A, Melino G, Candi E. Gene expression in organoids: an expanding horizon. Biol Direct. 2023;18(1):11. Published 2023 Mar 25. doi:10.1186/s13062-023-00360-2 Transcriptome analysis at the single-cell level determines the presence of the appropriate cell types in these complex 3D systems, confirming that engineered organoids are suitable for experimental models for native organs.5Smirnov A, Melino G, Candi E. Gene expression in organoids: an expanding horizon. Biol Direct. 2023;18(1):11. Published 2023 Mar 25. doi:10.1186/s13062-023-00360-2 Single-cell sequencing thus provides valuable assessment of cell type transcriptomics in these engineered tissues, even absence their spatial context.
A full understanding of various cell interactions in a tissue requires information about spatial biology.
A full understanding of various cell interactions in a tissue requires information about spatial biology, prompting the development of two single-cell sequencing techniques. One technique involves microscope slides pre-labelled with RNA probes for identifying the positions of the transcripts labeled in situ upon placement of the tissue section. The RNA is then released from the tissue, transcripts sequenced, and subsequent reconstruction of position information provides a two-dimensional map of tissue gene expression.6Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg Å, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, Lundeberg J, Frisén J. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016 Jul 1;353(6294):78-82. doi: 10.1126/science.aaf2403. PMID: 27365449.5Smirnov A, Melino G, Candi E. Gene expression in organoids: an expanding horizon. Biol Direct. 2023;18(1):11. Published 2023 Mar 25. doi:10.1186/s13062-023-00360-2 This approach requires several sample manipulations, including amplification and sequencing steps, that make spatial studies cumbersome and less robust than needed for clinical studies.
A separate technique uses the combined approach of single-cell sequencing and fluorescent in situ hybridization (FISH) to examine spatial context of gene expression.2Chan TW, Fu T, Bahn JH, Jun HI, Lee JH, Quinones-Valdez G, Cheng C, Xiao X. RNA editing in cancer impacts mRNA abundance in immune response pathways. Genome Biol. 2020 Oct 26;21(1):268. doi: 10.1186/s13059-020-02171-4. PMID: 33106178; PMCID: PMC7586670.7Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, Zhuang X. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018 Nov 16;362(6416):eaau5324. doi: 10.1126/science.aau5324. Epub 2018 Nov 1. PMID: 30385464; PMCID: PMC6482113. Although this technique provides a clearer picture of the spatial context of labeled transcripts, it also involves an amplification step and multiple sample manipulations. Even so, these single-cell sequencing techniques provide some spatial context for cellular gene expression profiles. However, newer technology by NanoString, discussed below, improves on this concept of in situ imaging and identification, removing the amplification steps and simplifying the workflow.
Newer techniques surpass single-cell sequencing
While single-cell sequencing has led to significant research advances, the ability to extensively profile gene expression rapidly and reliably in the spatial context of a tissue can accelerate those advances. Additionally, reliable techniques that are fast and easy to use are needed for clinical diagnosis of disease, a forum where spatial gene expression analysis is becoming an invaluable tool.
Improving on single-cell sequencing requires a paradigm shift away from direct sequencing techniques toward more information-rich and broadly applicable single-cell transcriptomic techniques.
Improving on single-cell sequencing requires a paradigm shift away from direct sequencing techniques toward the more information-rich and broadly applicable single-cell transcriptomic techniques. Known as spatial multiomics, several recently developed platforms localize and quantify gene expression profiles at the cellular and subcellular levels, including NanoString’s GeoMx DSP and CosMx SMI. Combining barcoded hybridization probes and quantification with immunofluorescence imaging, GeoMx DSP and CosMx SMI uncover the spatial relationships between different cell types in a tissue. Complementary to one another, GeoMx DSP can profile whole transcriptomes and the CosMx SMI can resolve gene expression profiles at the single-cell and subcellular levels, all in a spatial context perfect for mapping cell types and examining cell-cell interactions.

Since many human diseases have a molecular component, understanding which molecules are present at specific times and places can guide research toward new targets for potential therapies. Spatially mapping cellular organization and molecular distribution of cells and tissues using GeoMx DSP and CosMx SMI can uncover pertinent specialized roles of specific cells and molecules, identifying potential therapeutic targets.
GeoMx DSP Adds Spatial Context to Single-Cell Sequencing
The combination of 21st century digital technology, bioinformatics, and genomic sequence databases removes the need for conventional sequencing reactions to obtain transcriptomic data. Sophisticated hybridization protocols, using fluorescently-labeled barcoded probes, are replacing single-cell sequencing techniques. GeoMx DSP uses this technology to identify expressed transcripts in context of the tissue structure architecture without the PCR amplification steps needed in scRNA seq, decreasing the potential for sequencing errors.

Visualizing and quantifying RNA with GeoMx DSP gives spatial context for assessing interactions among cells at the tissue level. Providing a bird’s eye view of gene expression profiles in a tissue, GeoMx DSP can address biological questions such as inter- and intra-tumor heterogeneity, genomic instability, phenotypic variability, epigenetic/transcriptional/protein expression variability, and TME interactions. Unlike single-cell sequencing, GeoMx looks at the whole transcriptome with the ability to focus on the location of functional regions.
GeoMx DSP can assess all transcripts and proteins in their original location in architecturally-intact tissue samples.
Capable also of detecting protein expression, the multi-plex, high throughput environment of GeoMx DSP can easily analyze RNA and protein expression in the same tissue sample. With the sensitivity to detect high, medium and low expressing genes across a broad dynamic range, barcoded hybridization probes give researchers the power to perform “spatially-informed simultaneous assessment of multiple biomarkers.”8Toki MI, Merritt CR, Wong PF, Smithy JW, Kluger HM, Syrigos KN, Ong GT, Warren SE, Beechem JM, Rimm DL. High-Plex Predictive Marker Discovery for Melanoma Immunotherapy-Treated Patients Using Digital Spatial Profiling. Clin Cancer Res. 2019 Sep 15;25(18):5503-5512. doi: 10.1158/1078-0432.CCR-19-0104. Epub 2019 Jun 12. PMID: 31189645; PMCID: PMC6744974. GeoMx DSP can assess all transcripts and proteins in their original location in architecturally-intact tissue samples.9Vlachos I, What is Spatial Biology? Why is it the hottest field in Biomedicine? When will it transform Research and Clinical practice? How can I Enter?, 2022 https://www.linkedin.com/pulse/what-spatial-biology-why-hottest-field-biomedicine-when-vlachos
The power of GeoMx DSP is demonstrated by its use in over 200 publications, on topics ranging from cancer biology to immunology to neuroscience since its launch in 2019. GeoMx DSP has also been used to identify novel biomarkers, assess pertinent cell populations for understanding disease biology, and study the effects of candidate drugs. Combining Whole Transcriptome Atlas (WTA) analysis, multiplex runs, high throughput, and the ability to focus on functional regions, GeoMx DSP is an ideal choice for forward and reverse translational biomarker discovery studies that cast a wide net while using a minimal amount of limited samples. Furthermore, GeoMx DSP has been optimized for using archived FFPE tissue slides, giving researchers the power to access information from millions of patients across several decades. Add in its ability to analyze protein expression and GeoMx DSP has clearly surpassed single-cell sequencing for analyzing spatial multiomics.
How does CosMx SMI Differ from the GeoMx DSP?
Using technology similar to GeoMx DSP, CosMx SMI can resolve the fine details of gene expression in individual cells. The technology in GeoMx DSP is enhanced in CosMx SMI by employing multiple cycles of hybridization of fluorescent probes on intact tissue samples for high-resolution imaging and multi-analyte cell segmentation analysis. A proprietary deep-learning algorithm analyzes each cell boundary definition and subcellular resolution images overlaid on fluorescent expression probe images to provide precise cell segmentation in a three-dimensional reconstruction of the observed gene expression profile.10He, S., Bhatt, R., Brown, C. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-022-01483-z With this method, CosMx SMI is able to pinpoint the location of gene expression and molecular interactions in tissues and cells of up to 1,000 RNA transcripts and 64 proteins in a single experiment, making it the highest-plex multiomic imager available.
With enough sensitivity to detect low-expressing genes, CosMx SMI tracks gene expression changes and ligand-receptor interactions for classifying cell types, cellular functions and cell-cell interactions in a tissue to ensure no important biology is missed.10He, S., Bhatt, R., Brown, C. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-022-01483-z The first spatial multiomics platform optimized for formalin-fixed paraffin-embedded (FFPE) and fresh frozen (FF) tissue samples, CosMx SMI joins GeoMx DSP in replacing single-cell sequencing with more robust, spatially-defined gene expression profiling.
CosMx SMI joins GeoMx DSP in replacing single-cell sequencing with more robust, spatially-defined gene expression profiling.
The combination of GeoMx DSP’s high throughput, whole transcriptome analysis and CosMx SMI’s single-cell and subcellular resolution, high-plex RNA and protein provides spatialomics analysis for every level from tissue to subcellular for cell typing, microenvironment mapping, examination of ligand-receptor interactions, biomarker discovery, and comparison of diseased and healthy tissues.
What can spatial multiomics tell us about human disease?
Previous technologies, such as scRNA seq, provided important information about the transcriptomics of individual cells or subgroups of cells. However, scRNA seq misses the spatial relationship of cells to each other in a tissue’s architecture. The ability to characterize gene expression profiles within their cellular spatial relationships is critical to a fuller understanding of the biological function of tissues, particularly regarding changes occurring in a diseased tissue.
Spatial multiomics to characterize tissue microdomains
Supporting the importance of spatial biology is an example from patients infected with Human Immunodeficiency Virus (HIV). HIV is known to persist in viral reservoirs in cells and tissues after cessation of treatment. Dormant HIV is capable of reactivation, imposing on patients the cost and logistical challenge of long-term antiviral treatment.
Understanding the cellular biology of HIV viral reservoirs is therefore critical to relieving patients of this burden, and relies on the ability to locate and identify the reservoirs, and characterize the gene expression profiles of those cells. Spatial transcriptomics is a great technique for revealing this information. In a recent report from Plyler et al, lymph nodes from rhesus macaques were infected with an HIV-related virus, Simian Immunodeficiency Virus (SIV), and characterized with GeoMx DSP and CosMx SMI. It was found that tissue Regions of Interest (ROIs) with active viral infections showed significantly altered transcriptional profiles compared to uninfected ROIs. Molecular pathways affected in the infected ROIs included Il-4, Il-17, and HLA signaling, along with Interferon and inflammation pathways and B Cell Development. GeoMx spatial analysis discovered more than 200 genes showing differential expression, a result that would have been missed with pan-tissue or single-cell RNA sequencing approaches.11Plyler J, Kim S, Kim Y, Chan CN, Newhouse DJ, Gale MJ, Murphy S, Gregory M, Warren S, Vanschoiack A, Estes JD. Spatial Single Cell characterization of SIV reservoirs in lymphoid tissues and B cell follicles in rhesus macaques 2021 https://nanostring.com/wp-content/uploads/AMP-2021_SMI-0014_JacobEstes_OHSU_final.pdf
GeoMx analysis discovered more than 200 genes showing differential expression that would have been missed with single-cell RNA sequencing.
The resulting gene expression profile information was used to a develop a map of tissue microdomains, termed neighborhoods, for use in guiding CosMx SMI analysis. Localization was then fine-tuned for looking at single cells in each neighborhood to investigate differences in transcriptional dynamics between infected and uninfected cells and for identification of specific viral transcripts inside the cells. The resolution of CosMx SMI was capable of distinguishing between extracellular virions and infected cells, allowing researchers to further explore the mechanism of HIV viral reservoirs in hopes of discovering a method of eradicating them through improved therapies or potential cures.
Spatial multiomics in understanding intercellular communication
Proper function of biological systems requires cells to continually interact with one another. Important to maintaining tissue structure and function and overall system health, cells coordinate their activities via cell-cell interactions.
Cell-cell interactions involve several mechanisms. Stable cell-cell interactions include tight junctions and receptor protein-mediated cell signaling. Transient interactions include those found in immune cell migration to infection sites. Each type of cell interaction involves cellular communication, typically in response to changes in their microenvironment. Possibly the most critical cell-cell interaction for responding to changes is ligand-receptor interaction. Ligands, which can be membrane-bound or extracellular, bind to a specific receptor to signal the target cell, inducing molecular changes in that cell by setting off a signaling cascade. The induced subcellular changes are often modifications of the cell’s gene expression profile. The ability of GeoMx DSP and CosMx SMI to spatially localize and quantify these transcriptomic changes is valuable for examining the transcriptional dynamics of target cells.
A recent study used CosMx SMI to characterize ligand-receptor interactions of tumor cells and T cells by developing a spatial map of the cell types present and examining the interactions between the cells in tissue samples.10He, S., Bhatt, R., Brown, C. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-022-01483-z One hundred canonical ligand–receptor (LR) partners were identified, many of which were relevant to the tumor-immune interface, including immune checkpoints such as PD-L1/PD-1 and CTLA4/CD86.10He, S., Bhatt, R., Brown, C. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-022-01483-z Subsequent analysis of changes in these interactions across space and between samples was performed. Sixteen enriched LR pairs were discovered at the tumor–T cell interface in at least one of the five lung tumors examined.
The power of CosMx SMI to differentiate specific subcellular gene expression profiles was confirmed by its ability to show the presence of many interactions in only a subset of tumors.
The power of CosMx SMI to differentiate specific gene expression profiles at the subcellular level was confirmed by its ability to show the presence of many of these interactions in only a subset of tumors.10He, S., Bhatt, R., Brown, C. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-022-01483-z For instance, PD-L1/PD-1 (CD274/ PDCD1) exhibited a higher interaction score across four of the tumors but remained lower in the fifth one. Importantly, the variability in LR signaling strength observed between tumors was consistent with known variability in tumor response to immune checkpoint inhibitors and EGFR inhibitors.10He, S., Bhatt, R., Brown, C. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-022-01483-z CosMx Spatial Molecular Imager is therefore a valuable high throughput methodology for examining changes in gene expression in response to different cellular conditions.
What can spatial multiomics tell us about immunotherapy of cancer?
Even though the human body has a variety of fail-safe mechanisms to prevent cells from becoming malignant, just under two million people per year are diagnosed with cancer in the United States alone. Mechanisms for preventing malignancy include subcellular processes, such as cell-cycle checkpoints and apoptosis, and multicellular processes, such as those mediated by the immune system. Harnessing the immune system’s innate inclination to remove cancer cells requires understanding the molecular interactions between cancer cells and immune cells, which can be done by spatial multiomic analysis.
More accurate predictions of patient response to immunotherapies can be achieved by characterizing the spatial distribution of gene expression profiles in the tumor microenvironments.
Immune cells often interact with cancer cells when tumor infiltrating lymphocytes (TILs) invade a tumor. A recent study used CosMx SMI to profile TILs in renal cell carcinoma tissues, resulting in the identification of distinct microenvironments within the tumor tissue.12Newell EW, Kim Y, Ryu H, Li S, Simoni Y, Leon M, Kim S, Gregory M, Danaher P, Warren S, Beechem J. In-situ visualization and measurement of tumor-infiltrating lymphocytes (TILs) on intact FFPE renal cell carcinoma (RCC) tissue using the spatial molecular imager (SMI) 2021 https://nanostring.com/wp-content/uploads/SITC2021-poster_SMI-0001_EvanNewell_Hutch.pdf A subset of TILs, identified as T cells, exhibited a specific gene expression signature and were more highly enriched in tumor tissue regions compared to non-tumor regions. Since the phenotypes of TILs in cancer patients tend to be highly heterogeneous, predicting their response to immunotherapy treatment, designed to restore anti-tumor T cell-mediated immunity, is rather difficult. More accurate predictions of patient response to immunotherapies can be achieved through a better understanding of the role of TILs by characterizing the spatial distribution of gene expression profiles in the tumor microenvironments (TME). Additionally, such characterization may lead to the development of better immunotherapy solutions.12Newell EW, Kim Y, Ryu H, Li S, Simoni Y, Leon M, Kim S, Gregory M, Danaher P, Warren S, Beechem J. In-situ visualization and measurement of tumor-infiltrating lymphocytes (TILs) on intact FFPE renal cell carcinoma (RCC) tissue using the spatial molecular imager (SMI) 2021 https://nanostring.com/wp-content/uploads/SITC2021-poster_SMI-0001_EvanNewell_Hutch.pdf
In another study, Grice and colleagues used GeoMx DSP and CosMx SMI to create a spatial atlas for three types of skin cancer (basal cell carcinoma, squamous cell carcinoma, and melanoma). The atlas was then used to characterize cell-cell interactions involving PD-1+ immune cells and PD-L1+ cancer cells specifically at the tumor boundary. Characterizing the specifics of these interactions will increase understanding of each type of skin cancer is caused, potentially leading to more effective treatments.13Grice LF, Jin X, Tran M, Mulay O, Killingbeck E, Gregory M, Teoh SM, Devitt K, Kulasinghe A, Leon M, Murphy S, Warren S, Khosrotehrani K, Stark M, Frazer I, Kim Y, Nguyen Q. Building a spatial single-cell multi-omics atlas and cellular interactome for skin cancer. 2021 https://nanostring.com/wp-content/uploads/HCA2021_final.pdf
The promise of examining multiomics with spatial biology analysis
Proper functioning of biological systems revolves around molecular biology processes occurring in the right place at the right time in the appropriate amount. Spatial biology technologies such as those utilized by GeoMx DSP and CosMx SMI provide unprecedented insights into the molecular biology of cells, enhancing understanding of how disease states arise and guiding research toward potential treatments. Spatial biology techniques such as these surpass single-cell sequencing by providing the spatial information to give specific molecular context for biological processes of interest. This technology will continue to open new doors for research into the origins and treatments of disease processes.
The CosMx® SMI and decoder probes are not offered and/or delivered to the Federal Republic of Germany for use in the Federal Republic of Germany for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof in a method used in fluorescence in situ hybridization for detecting a plurality of analytes in a sample without the consent of the President and Fellows of Harvard College (Harvard Corporation) as owner of the German part of EP 2 794 928 B1. The use for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof is prohibited without the consent of the President and Fellows of Harvard College (Harvard Corporation).
References
- 1Li Y, Jiang X, Zhong M, Yu B, Yuan H. Whole Genome Sequencing of Single-Circulating Tumor Cell Ameliorates Unraveling Breast Cancer Heterogeneity. Breast Cancer (Dove Med Press). 2022 Dec 28;14:505-513. doi: 10.2147/BCTT.S388653. PMID: 36597488; PMCID: PMC9805725.
- 2Chan TW, Fu T, Bahn JH, Jun HI, Lee JH, Quinones-Valdez G, Cheng C, Xiao X. RNA editing in cancer impacts mRNA abundance in immune response pathways. Genome Biol. 2020 Oct 26;21(1):268. doi: 10.1186/s13059-020-02171-4. PMID: 33106178; PMCID: PMC7586670.
- 3Grun D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525(7568):251–5.
- 4Kumar MP, Du J, Lagoudas G, Jiao Y, Sawyer A, Drummond DC, Lauffenburger DA, Raue A. Analysis of Single-Cell RNA-Seq Identifies Cell-Cell Communication Associated with Tumor Characteristics. Cell Rep. 2018 Nov 6;25(6):1458-1468.e4. doi: 10.1016/j.celrep.2018.10.047. PMID: 30404002; PMCID: PMC7009724.
- 5Smirnov A, Melino G, Candi E. Gene expression in organoids: an expanding horizon. Biol Direct. 2023;18(1):11. Published 2023 Mar 25. doi:10.1186/s13062-023-00360-2
- 6Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg Å, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, Lundeberg J, Frisén J. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016 Jul 1;353(6294):78-82. doi: 10.1126/science.aaf2403. PMID: 27365449.
- 7Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, Zhuang X. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018 Nov 16;362(6416):eaau5324. doi: 10.1126/science.aau5324. Epub 2018 Nov 1. PMID: 30385464; PMCID: PMC6482113.
- 8Toki MI, Merritt CR, Wong PF, Smithy JW, Kluger HM, Syrigos KN, Ong GT, Warren SE, Beechem JM, Rimm DL. High-Plex Predictive Marker Discovery for Melanoma Immunotherapy-Treated Patients Using Digital Spatial Profiling. Clin Cancer Res. 2019 Sep 15;25(18):5503-5512. doi: 10.1158/1078-0432.CCR-19-0104. Epub 2019 Jun 12. PMID: 31189645; PMCID: PMC6744974.
- 9Vlachos I, What is Spatial Biology? Why is it the hottest field in Biomedicine? When will it transform Research and Clinical practice? How can I Enter?, 2022 https://www.linkedin.com/pulse/what-spatial-biology-why-hottest-field-biomedicine-when-vlachos
- 10He, S., Bhatt, R., Brown, C. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-022-01483-z
- 11Plyler J, Kim S, Kim Y, Chan CN, Newhouse DJ, Gale MJ, Murphy S, Gregory M, Warren S, Vanschoiack A, Estes JD. Spatial Single Cell characterization of SIV reservoirs in lymphoid tissues and B cell follicles in rhesus macaques 2021 https://nanostring.com/wp-content/uploads/AMP-2021_SMI-0014_JacobEstes_OHSU_final.pdf
- 12Newell EW, Kim Y, Ryu H, Li S, Simoni Y, Leon M, Kim S, Gregory M, Danaher P, Warren S, Beechem J. In-situ visualization and measurement of tumor-infiltrating lymphocytes (TILs) on intact FFPE renal cell carcinoma (RCC) tissue using the spatial molecular imager (SMI) 2021 https://nanostring.com/wp-content/uploads/SITC2021-poster_SMI-0001_EvanNewell_Hutch.pdf
- 13Grice LF, Jin X, Tran M, Mulay O, Killingbeck E, Gregory M, Teoh SM, Devitt K, Kulasinghe A, Leon M, Murphy S, Warren S, Khosrotehrani K, Stark M, Frazer I, Kim Y, Nguyen Q. Building a spatial single-cell multi-omics atlas and cellular interactome for skin cancer. 2021 https://nanostring.com/wp-content/uploads/HCA2021_final.pdf
 The CosMx® Spatial Molecular Imager
The CosMx® Spatial Molecular Imager