Spatial Multiomics Single-Cell Imaging Platform to Explore FFPE Tissues with Spatial Context
What is Spatial Multiomics?
Spatial multiomics describe the ability to locate and quantify DNA, RNA, and protein in the three-dimensional space of a tissue. Decades of research have built a solid knowledge base for these macromolecules, their biochemistry, molecular interactions, and expression patterns. It was developed to characterize these macromolecules, and their expression profiles, in the intricate three-dimensional architecture of the tissue to exhaustively understand their roles in biological functions.
To accomplish this, a tissue of interest is subjected to a panel of transcripts and/or proteins labeled with fluorescent probes. After visualizing regions of interest (ROI), the transcripts or proteins in those ROI are identified and counted. Tabulated numbers are then related back to the ROI, elucidating the gene expression profiles at play in the tissue environment. This process requires a high plex and data analysis capacity, which traditional methods lacked.
Combining current computing and imaging technologies with the extensive genome sequence information available brings biology closer to the Holy Grail of quantifying gene expression in three-dimensional space. Analyzing genomics, transcriptomics, and proteomics, singly or in combination, in the 3D space of a tissue constitutes a new field known as spatial biology multiomics or spatial multiomics.
In this blog, we will explore the possibilities unlocked by the application of high plex single-cell imaging and transcriptomics research.
Spatial Transcriptomics: from Bulk to Clear Picture
The technology has quickly gained acceptance as the premier technology for studying molecular and cellular interactions. Designed to give a clear picture of the inter- and intra-cellular molecular dynamics at play in a tissue environment, spatial transcriptomics combines quantitative capabilities of bulk expression systems (such as the nCounter® Pro) with fluorescent imaging to visualize and quantify gene expression.
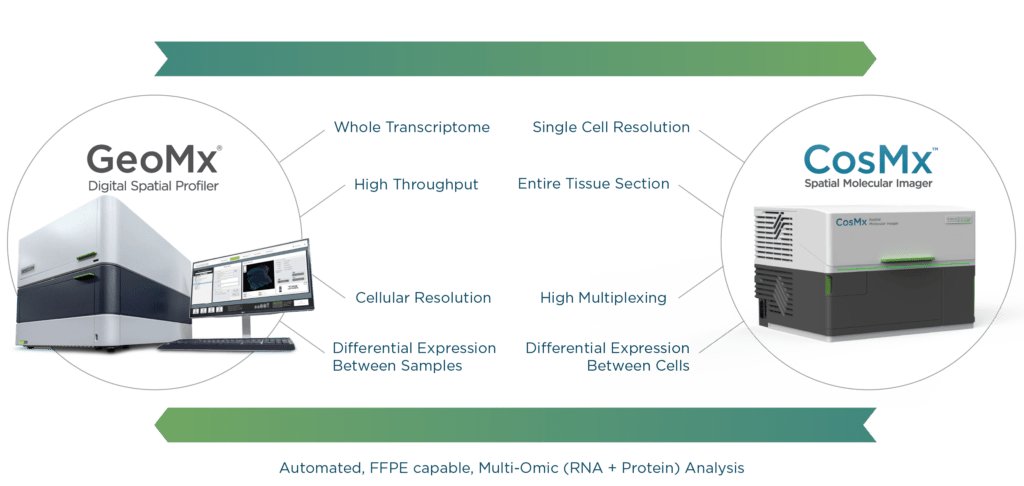
Indeed, industry-leader NanoString Technologies introduced the GeoMx® Digital Spatial Profiler (DSP) in 2019, an automated system designed specifically for histological slides from formalin-fixed paraffin-embedded (FFPE) tissue samples. Adapting our proven molecular barcode technology, we felt it was imperative, given the prevalence of FFPE samples in labs and clinics, that researchers have a quick and reliable method for extracting expression data from their banks of tissues.
Having already established the nCounter bulk expression platform as the best in class for FFPE materials in quantifying RNA expression profiles (in addition to other sample types), it made sense to modify that technology for a spatial transcriptomics platform.
What is Spatial Transcriptomics Used For?
Spatial transcriptomics is designed to derive more gene expression profiling information from tissue slides to untangle the complexity of biological systems at the cellular and subcellular levels. Going beyond merely visualizing the expression patterns of a few genes in a tissue, spatial omics can visualize and quantify hundreds of genes at once. By quantifying expression profiles in three-dimensional space, spatial omics are leading to a greater understanding of the regulation of genes of interest and their downstream effects within cells and tissues. Every field of biology, from development to differentiation to disease processes, can benefit from the precision spatial omics bring to tracking changes in gene expression profiles in a cellular neighborhood. Comparing gene expression profiles under various conditions in a spatial context can help us better understand how to manipulate biological systems.
Spatial Transcriptomics and Gene Expression Profiles as Biomarkers
Probably the most valuable use of spatial transcriptomics is following gene expression profiles as biomarkers. Its ability to provide a “spatially informed simultaneous assessment of multiple biomarkers (Toki et al.)” has been used in several studies to date. For example, it has been used to characterize immune cell infiltration in kidney disease anti-neutrophil cytoplasmic autoantibody-associated glomerulonephritis (1).
Frequently progressing to end-stage kidney disease, little is known about the pathogenesis of ANCA-GN. However, it was known that glomerular injury in ANCA-GN involves the loss of podocytes, cells critical for tissue architecture and function. Located in a special environment, podocytes are separated from outside tissues by the Bowman’s capsule (BC) and from the capillary lumen by the glomerular basement membrane (1). Previous studies showed that the architecture of this special environment protects podocytes from an influx of cytotoxic CD8+ T cells in normal tissue.
Using digital spatial profiling, Ye et al. were able to identify immune cell dynamics that indicated disease progression. Ye concluded their results show “that digital spatial profiling allows the comparative analysis of the mRNA and protein profiles in individual glomeruli and the identification of potential novel mechanisms in ANCA-GN (1).” Visualization and quantitation of gene expression profiles using spatial omics thus led them to a better understanding of the disease.
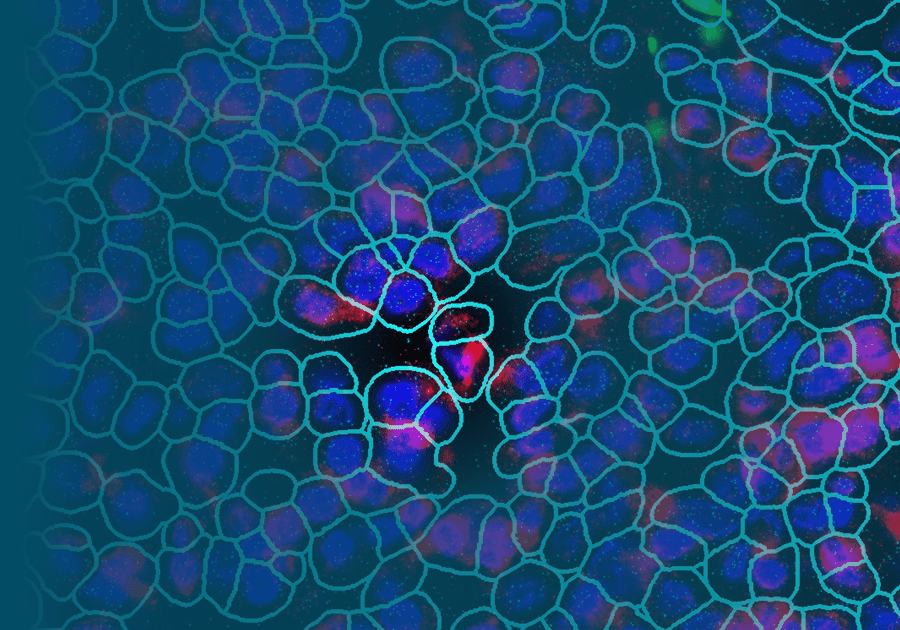
Spatial Transcriptomics and Single-Cell Imaging
While Ye et al. used a spatial biology profiling system of their own, similar work was done in kidneys by Newell et al. to study renal cell carcinoma (RCC) using the CosMx™ Spatial Molecular Imager (SMI) for single-cell imaging system from NanoString® (2). They characterized the heterogeneous population of tumor-infiltrating lymphocytes (TIL) to further understand the tumor microenvironment (TME) that varies from patient to patient.
Not only did they find virus-specific bystander T cells in RCC tumors with diverse phenotypic profiles, but they also identified distinct niches (e.g., tumor-rich, stroma, and distinct regions with high densities of immune cell infiltrates such as ones co-enriched for T and B cells) within the TME of RCC tumor sections. SMI analysis of RCC, mirroring studies done in other tissues, and clearly defined subsets of cells with similar gene expression profiles to further our understanding of the TME (2).
Single-Cell Imaging and CRISPR
In addition to comparing normal and diseased tissue environments, spatial transcriptomics can also be used in combination with CRISPR to examine the effects on gene expression profiles in response to genetic manipulation by CRISPR (3). In their hands, the SMI assay was capable of simultaneously detecting guide RNAs (gRNAs, used for directing CRISPR to the desired genomic sequence) and quantifying RNA expression of genes of interest in the same cell.
With the ability to scale the plex of the target screen up to 1000 genes while interrogating >100,000 cells, Freedman used SMI in conjunction with CRISPR on the LNCaP prostate cancer cell line to apply epigenetic perturbations to an androgen receptor (AR) enhancer. SMI analysis validated the impact of the introduced modifications on the AR gene and its downstream targets. Furthermore, Freedman showed that the SMI system has a high resolution, giving unambiguous visualization and digital detection of gRNAs in single cells while also demonstrating high sensitivity by detecting key low expression (TPM ~ 1-10) AR-related target genes (3).
Additionally, quantitation of AR-related target genes was observed on over 90% of the cells analyzed, with a high detection sensitivity of rare, modified cells (below 1%). The high plex capability and flexible panel design means it is possible to visualize and measure up to 1000 gRNAs and genes at single-cell resolution. Combined with accurate quantitation and a simple high-throughput workflow, spatial omics analysis, and CRISPR technology will give researchers the power to study the downstream effects of any genomic modifications.
Single-Cell Imaging Supports Spatial Transcriptomics in Studying Archived FFPE Samples
Spatial transcriptomics can also be used to add information to previous studies with the addition of Single-Cell Imaging. Having been designed specifically for FFPE tissue samples, GeoMx® Digital Spatial Profiler and CosMx™ Spatial Molecular Imager can add spatial biology analysis to previous studies using archived FFPE samples. Indeed, for cancer research, this can be a big advantage. As He et al. (4) wrote, “Real-world FFPE cancer samples are challenging to image at high-plex in a multi-omic manner using conventional methods.”
GeoMx and CosMx bring high-plex and multi-omic capabilities to FFPE samples, while harnessing the advantages they have over other samples such as fresh frozen tissue, including (1) preservation of cellular and architectural morphology; (2) the ability to store at room temperature for several years; and (3) easy availability, as they are routinely prepared in labs and clinics. The GeoMx DSP can visualize RNA transcripts from up to 22,000 genes, or up to 150 proteins, on one slide at one time, while CosMx SMI can visualize up to 1,000 RNA transcripts and 100 validated proteins within a single cell. Both systems can perform automated quantitation of biomarkers within selected ROI. Insights into gene expression profiles at the cellular and subcellular levels can thus be added to the knowledge base for a given cancer or tissue.
What is Single-Cell Imaging Spatial Transcriptomic, and How is it Different from Single-Cell Gene Expression?
Single-cell imaging spatial transcriptomics is a more information-laden version of single-cell gene expression. In single-cell gene expression, the desired cell type is isolated from a tissue for analysis of the specific genes being expressed in it. While the cell-type expression is valuable, separating the cell from its structural neighborhood means a loss of 3D information. Being able to preserve the tissue architecture while examining gene expression provides clues as to how the cellular environment functions. This is where single-cell imaging spatial transcriptomics delivers on the promises of present-day molecular, cellular, genetic, histological, and computer technology.
GeoMx Digital Spatial Profiler and CosMx Spatial Molecular Imager are both capable of quantifying gene expression profiles in the context of tissue architecture. The difference comes down to resolution. GeoMx DSP is higher throughput but with slightly lower resolution, while CosMx SMI gives up some throughput capacity in favor of higher resolution. GeoMx has the optics to enable single-cell capture, but CosMx SMI is an in-situ technology designed for single-cell measurement. Its sensitivity is tuned to the single cell, whereas GeoMx DSP aims for groups of cells (ROIs or AOIs). In-situ detection in CosMx SMI improves information capture from a single cell, unlike the release-and-read approach used in GeoMx DSP.
Furthermore, CosMx SMI has a small readout zone that uses segmentation markers and algorithms to enable individual visualization of each cell, not just ones well-discriminated by morphology markers (Erin Piazza, NanoString). Both GeoMx DSP and CosMx SMI employ probes that are easily adapted to oligo-labeled antibodies, which can be used in concert on the same tissue slides to correlate RNA and protein expression in a tissue. These advances in spatial transcriptomic technology are what truly set GeoMx DSP and CosMx SMI ahead of other spatial transcriptomic methods.
Summary
Ever since biological macromolecules were first described, biologists have been chasing the dream of examining and understanding DNA, RNA, and protein functions at the cellular and molecular levels. With spatial transcriptomics, all the advances in biochemistry, molecular biology, microscopic imaging, and bioinformatics from previous decades come together to bring biology closer than ever to achieving that dream.
The confluence of technology present in GeoMx Digital Spatial Profiler and CosMx Spatial Molecular Imager gives researchers the power to spatially resolve biomarkers within the cellular mosaic of tissue and define functional segmentation within that tissue. Understanding tissue heterogeneity is crucial to studying developmental biology, disease pathogenesis, and response to treatment. Being able to analyze the spatial distribution of gene expression profiles in a tissue is therefore fundamental to a full understanding of gene function. A machine that can do so in an efficient and reliable manner is an invaluable research tool.
For Research Use Only. Not for use in diagnostic procedures.
The CosMx™ SMI and decoder probes are not offered and/or delivered to the Federal Republic of Germany for use in the Federal Republic of Germany for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof in a method used in fluorescence in situ hybridization for detecting a plurality of analytes in a sample without the consent of the President and Fellows of Harvard College (Harvard Corporation) as owner of the German part of EP 2 794 928 B1. The use for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof is prohibited without the consent of the President and Fellows of Harvard College (Harvard Corporation).
References:
- Lin Ye, Yu Liu Xuejing Zhu, Tongyue Duan, Chang Wang, Xiao Fu, Panai Song, Shuguang Yuan, Hong Liu, Lin Sun, Fuyou Liu, Kyung Lee, John Cijiang He, Anqun Chen Digital Spatial Profiling of Individual Glomeruli From Patients With Anti-Neutrophil Cytoplasmic Autoantibody-Associated Glomerulonephritis Front Immunol 2022 Mar 2;13:831253. doi: 10.3389/fimmu.2022.831253. eCollection 2022.
- Evan W. Newell, Youngmi Kim, Heeju Ryu, Shamin Li, Yannick Simoni, Michael Leon, Sean Kim, Mark Gregory, Patrick Danaher, Sarah Warren, Joseph Beechem In-situ visualization and measurement of tumor-infiltrating lymphocytes (TILs) on intact FFPE renal cell carcinoma (RCC) tissue using the spatial molecular imager (SMI) 2021 AGBT Poster.
- Matthew L. Freedman, Sandor Spisak, Ji-Heui Seo, Talal El Zarif, Shanshan He, Joe Phan, Isabel Lee, Justin Jenkins, Erin Piazza, Dae Kim, Joseph Beechem Highly Sensitive transcriptomic-based pooled CRISPR screening enabled by Spatial Molecular Imager (SMI)” 2021 AGBT poster.
- He et al., 2021 High-Plex Multiomic Analysis in FFPE Tissue at Single-Cellular and Subcellular Resolution by Spatial Molecular Imaging.

