GeoMx® DSP Spatial Proteogenomics datasets now available on Illumina® BaseSpace™ Sequence Hub
NanoString and Illumina have collaborated to release two new demo datasets of GeoMx Digital Spatial Profiler (DSP) data on the Illumina BaseSpace Sequence Hub (BSSH) cloud.
Spatial proteogenomics enables new capabilities that use sequencing-based readout to combine RNA and protein expression analysis in FFPE tissue and fresh frozen tissue sections with spatial location. The GeoMx DSP platform enables spatial proteogenomics workflows to gain rich proteomic and whole transcriptome datasets from the same tissue section. Illumina’s BaseSpace Sequence Hub (BSSH) is a cloud-based genomics hub for NGS data management and analysis. Together, we’ve paired GeoMx DSP with NGS readout on Illumina sequencing instruments to enable spatial, multiomic scalability from your samples of interest.
The spatial proteogenomics workflow

Together, we have previously released a DRAGEN™ accelerated app hosted on BSSH, called the GeoMx NGS Pipeline. This app works to analyze and format sequencing data for compatibility with GeoMx DSP. Further, we’ve released integrated end-to-end workflows that streamline Illumina sequencer run planning from the GeoMx DSP instrument with the options for on-board or auto-launched BSSH analysis using the GeoMx NGS Pipeline app with a NextSeq 1000/2000 run.
Today, we’re pleased to announce the publication on BSSH of a new demo dataset of GeoMx DSP data on glioblastoma spatial proteogenomics as well as one on mouse embryonic development. Each dataset was previously run in-house at NanoString, uploaded to BSSH, and re-analyzed using the BSSH GeoMx NGS Pipeline app. Below are links to directly import the runs and project folders into your BaseSpace account. These runs can be found under the “Spatial omics” and “Multiomics” categories. Because the datasets are public, they are free, and do not count against storage limits. See Illumina’s blog post to understand how to access these data.
You can use the demo data to compare with your own spatial transcriptomics/proteogenomics runs. See the first Illumina blog post in this series for additional details on how to evaluate your sequencing run quality.
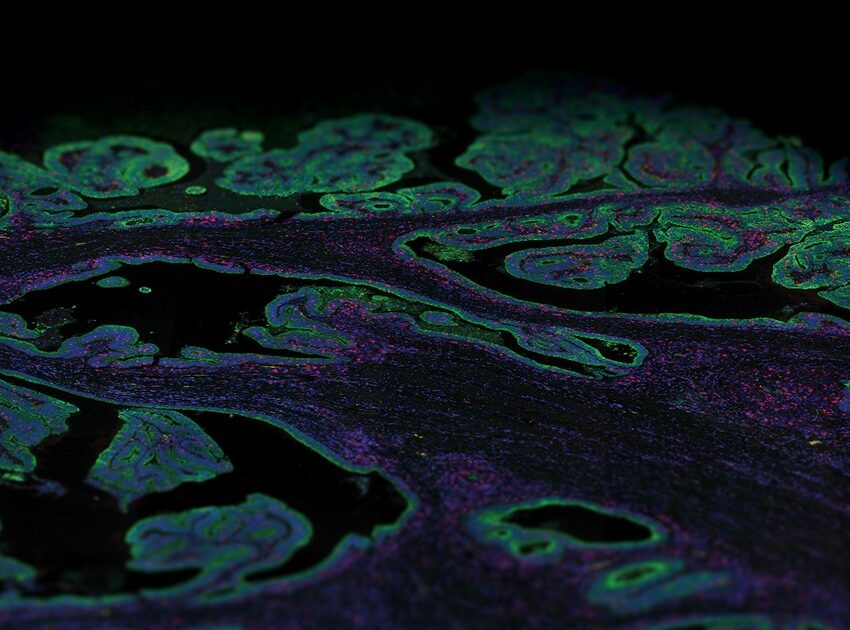
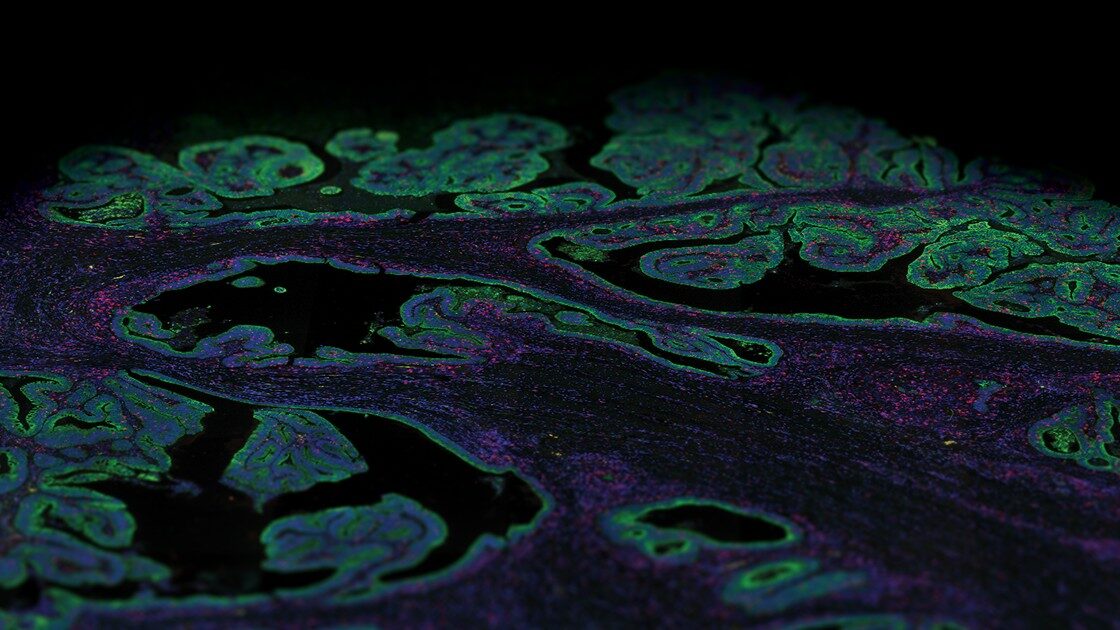
The spatial proteogenomics dataset collected on the NovaSeq 6000 features 306 libraries from a human glioblastoma multiforme tissue microarray. Samples were profiled both with the GeoMx® Human Whole Transcriptome Atlas (18,000+ targets) and 15 protein panels (100+ targets). This dataset is used in the GeoMx Spatial Proteogenomics publication. The other project run with the NextSeq 2000 has 357 libraries from six fixed frozen tissue sections derived from one E13 C57BL/6 mouse embryo and were profiled with the GeoMx® Mouse Whole Transcriptome Atlas (20,000+ targets). These public datasets are part of our Spatial Organ Atlas and the WTA publication.
The mouse development samples are part of our Spatial Organ Atlas, and are also featured in the 2022 Genome Research publication and are available via GEO (GSE190089). We hope these demo datasets will be highly useful to share with potential GeoMx customers.
For assistance with the GeoMx assay or data analysis with the GeoMx NGS Pipeline BaseSpace App, contact support@nanostring.com. For answers to sequencing questions, contact techsupport@illumina.com. To learn more about NextSeq 1000/2000 and the NovaSeq 6000, please visit the Illumina website.
BSSH demo data for project 1: Spatial Proteogenomics
NovaSeq 6000: NanoString GeoMx® Digital Spatial Profiler Spatial Proteogenomics on S4 35 cycles kit
30 tissues from 57 cores of human glioblastoma multiforme, totaling 302 regions and 4 NTCs, were profiled with NanoString GeoMx® Digital Spatial Profiler (DSP)’s RNA Whole Transcriptome Atlas (~19k targets) and 15 protein panels (~150 targets). Libraries were prepared with NanoString Seq Code primers and dual-index workflow (8 bp). Paired-end 2×27 sequencing was performed with a NovaSeq 6000 using an S4 35 cycle v1.5 kit. Loading concentration was 250 pM. Control PhiX was 1%. Gene expression was quantified using the GeoMx NGS Pipeline BaseSpace Sequence Hub app. These samples are featured in the GeoMx DSP ultra-high-plex Spatial Proteogenomics publication.
More on Illumina BaseSpace (requires login):
BSSH demo data for project 2: Mouse Development
NextSeq 2000: NanoString GeoMx® Digital Spatial Profiler Spatial Organ Atlas Mouse development on P3 50 cycles kit
351 profiled regions + 6 NTCs from six E13 C57BL/6 fresh frozen mouse embryos were profiled with the DSP’s RNA Mouse Whole Transcriptome Atlas (WTA) (~20k targets). Libraries were prepared with NanoString Seq Code primers and dual-index workflow (8 bp). Paired-end 2×27 sequencing was performed with a NextSeq 2000 using a P3 50 cycle kit. Loading concentration was 650 pM. Control PhiX was 5%. Gene expression was quantified using the GeoMx NGS Pipeline BaseSpace Sequence Hub app. These samples are part of NanoString’s Spatial Organ Atlas and the WTA publication.
More on Illumina BaseSpace (requires login):
Additional Resources
- Joint Application Note: High-resolution, high-throughput spatial transcriptomics of complex tissues [PDF]
- Joint Application Note: High-plex spatial proteogenomics of FFPE tissue sections [PDF]
- Original Workflows Blog Post: NanoString GeoMx® Digital Spatial Profiling with DRAGEN on BaseSpace™ Sequence Hub: Scaling NanoString spatial genomics with Illumina Sequencing
- E2E Workflow Blog Post: GeoMx® DSP Integrated with Sequencing Run Planning for NextSeq 1000/2000
- News post about NSTG at AGBT 2022, our partnership together: The spatial multiomics revolution
- Other BSSH demo dataset project blurbs: Illumina Demo Data