Common questions in molecular biology: Why is it called DNA barcoding?
DNA barcoding was named for its conceptual similarity to the Universal Product Codes (UPCs) used by supermarkets and other retailers to distinguish commercial products via a unique digital code. Based on the discovery of a short, standardized genetic region found to have unique DNA sequence in each species of fish, DNA barcoding was proposed as a system for species identification (Hebert). The concept of using unique DNA sequences for barcoding purposes has since been adopted by biologists for a variety of uses, including gene expression studies.
Naturally-occurring barcodes
DNA barcodes used for species classification are “scanned” by sequencing the DNA of the appropriate genomic region, analogous to the digital scanner used in supermarkets for identifying items via their barcode. Taxonomic identification of a specimen by DNA barcoding is performed by sequencing a short fragment (a few hundred base pairs) of the reference gene. The initial genomic region designated for classification purposes was the mitochondrial Cytochrome C Oxidase I (COI) subunit gene, which is conserved across species but contains species-specific sequence useful for identifying an unknown specimen. A 648 bp region of the COI gene from the unidentified specimen is amplified by PCR and sequenced for comparison to a database of known taxonomic barcodes (Hebert).
DNA barcoding accelerates species identification and provides better information about biodiversity
The uniqueness of the DNA barcode regions accelerates species identification and provides better information about biodiversity (Miller 2007). In fact, subsequent genomic regions in other phyla have been identified for use as DNA barcodes, including the internal transcribed spacer rRNA (ITS1-2) regions in fungi (Druzhinina), the ribulose-bisphosphate carboxylase (rbcL) and maturase K (matK) region in plants, and the 16S rRNA in prokaryotes (McGee). These naturally-occurring DNA barcodes allow users to efficiently categorize unidentified individuals using information from small genomic regions, a method which, theoretically, can be applied to all species of life (Kress).
Engineered DNA barcodes
The discovery of unique naturally-occurring DNA sequences that serve as barcodes inspired the idea to engineer unique DNA sequences for use as an identification tool. Engineered DNA barcodes are now being used as unique identifiers in various biological systems, further extending the similarity with UPC barcode generation.
Applications using unique DNA (or RNA) sequences as barcodes include signature-tagged mutagenesis (Mazurkiewicz), multiplexed high-throughput pyrosequencing (Parameswaran), and identifying insertions in viral vectors (Chen). These barcodes are identified via PCR or sequencing to confirm their presence in the molecules of interest. The heritability of DNA barcodes enables their use in tracking cell lineages within a population over multiple generations. Known as barcode lineage tracking (BLT), introduction through recombinant DNA techniques of barcodes facilitates characterization of T-cell recruitment, tracing cellular differentiation during organismal development, studying the clonal history of metastasis in cancer, screening and characterizing mutant libraries, and studying evolutionary dynamics (Johnson).
Essentially, any technique using recombinant DNA can use engineered DNA barcodes as unique identifiers.
DNA barcode probes
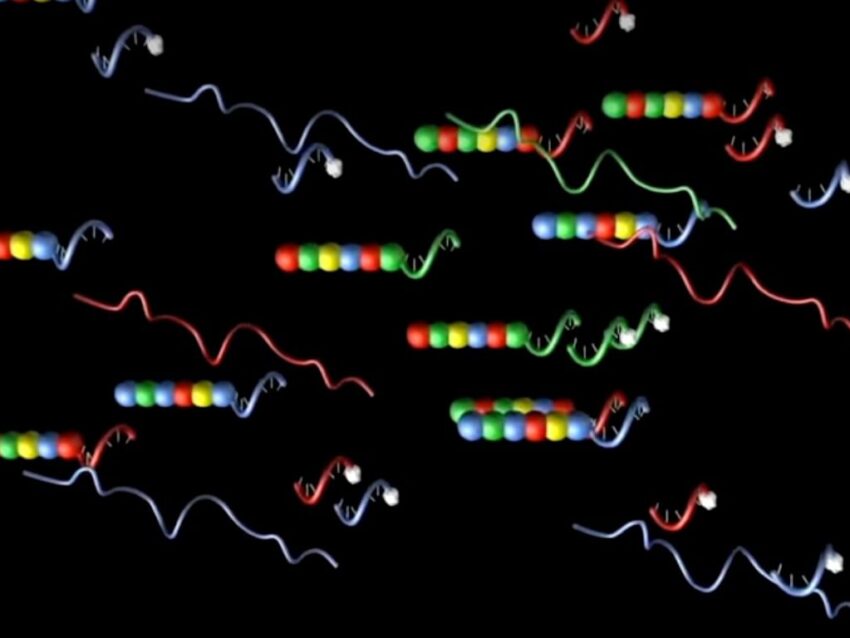
More recently, DNA barcodes have been adapted as probes, incorporating fluorescent molecules for easy identification of RNA transcripts during gene expression studies. Unique DNA sequences complementary to fluorescently-labeled RNA sequences are the basis of the barcode technology employed by NanoString in the nCounter® Pro Analysis System. The nCounter Pro screens tissue RNA transcripts by using panels of specific DNA sequences conjugated to unique fluorescently labeled RNA barcodes.
Unique DNA sequences complementary to fluorescently-labeled RNA sequences are the basis of the barcode technology employed by nCounter Pro.
The unique fluorescent color schemes provide a simple and efficient method of gene expression profiling in tissue samples (Geiss). It is also one of the first examples of barcoded probes that incorporate multiple types of molecules and is more accurately called a molecular probe. Whether the unique identifier sequence is composed of DNA or RNA, the concept of using a known unique sequence of molecules is the key element of any DNA or molecular barcode. Molecular barcoding applications will continue to expand, advancing research in many ways.
References
Béné MC, Lacombe F, Porwit A. Unsupervised flow cytometry analysis in hematological malignancies: A new paradigm. Int J Lab Hematol. 2021 Jul;43 Suppl 1:54-64. doi: 10.1111/ijlh.13548. PMID: 34288436.
Silveira GF, J Evolution of Flow Cytometry Technology Microb Biochem Technol 2015, 7(4):213-6 DOI: 10.4172/1948-5948.1000208

Related Content