How DNA Barcoding Improves Gene Expression Analysis and Biomarker Discovery
The phrase “DNA barcoding” can be applied to several methods of using unique DNA sequences for identification purposes. Sometimes known as “molecular barcoding,” DNA barcoding describes the use of unique naturally occurring sequences, as well as engineered sequences, for identification purposes. DNA barcodes are used in various techniques for a variety of reasons, including the classification of organisms into species groups, identifying individual cell types in a tissue, and transcriptomic analysis. All of these uses rely on the unique sequences in DNA barcodes to identify the biological entity of interest.
The discovery of DNA barcodes
One of the first uses of DNA barcoding was the discovery of unique genomic sequences that could distinguish different species from each other (Hebert). Taxonomy biologists used the mitochondrial gene cytochrome C oxidase I (COI) from a wide range of species to develop reference sequence libraries to serve as the core of a global bioidentification system for animals. DNA sequences unique to each species were identified from a signature region of the genome, for the purpose of assigning individuals in the wild to the appropriate species based on these sequence libraries (Hebert). All sequences used in this version of DNA barcoding are naturally occurring, discovered from available genomic sequence data for inclusion in the species identification reference database, though their uniqueness makes them suitable for other identification purposes.
Engineering DNA barcodes for transcriptomic analysis
Naturally occurring DNA barcodes inspired the idea to design and incorporate unique sequences into experimentally relevant DNA as molecular tags for cellular and molecular identification purposes.
Early uses of lab-derived DNA barcodes include signature-tagged mutagenesis (Mazurkiewicz), multiplexed high-throughput pyrosequencing (Parameswaran), and identifying insertions in viral vectors (Chen). Each of these uses involves the inclusion of DNA barcodes in a sequence of interest. For instance, in signature-tagged mutagenesis, barcode sequences are inserts into mutant libraries to reliably identify mutants from pools of DNAs in high-throughput functional screens of entire genomes (Mazurkiewicz).
The inclusion of DNA barcodes in cDNAs constructed from RNA libraries is designed to substantially enhance multiplexed high-throughput pyrosequencing by allowing pooling of DNA from independent samples and subsequent segregation to overcome errors typical to this method (Parameswaran). DNA barcodes included in a viral vector used to construct a library of insertional mutations provides a means of detecting and identifying the insertions in phenotypically mutant yeast (Chen). Each of these examples use unique DNA barcodes as means of identifying sequences of interest from a pool of DNAs, whether genomic or cDNA.
DNA barcode technology is now more accurately known as molecular barcoding, since the concept of using unique sequences for identification purposes is being applied to biomolecules other than DNA.
DNA barcode technology is now more accurately known as molecular barcoding, since the concept of using unique sequences for identification purposes has been applied to additional biomolecules. Various combinations of DNA, RNA, and protein are used to form hybrid reporter molecules, with the barcode portion residing in the nucleic acid component. Sequences used for these barcodes are often engineered, though naturally occurring sequences can still be used for some applications.
For instance, oligonucleotide-barcoded antibodies were used to characterize extracellular vesicles by their inner and outer proteins (Martel). Using engineered DNA sequences in combination with other biomolecules such as proteins provides flexibility for use of molecular barcodes in a wide variety of applications.
How molecular barcodes help accelerate GEP analysis
Molecular barcodes are proving to be nearly ideal for screening gene expression profiles (GEPs) of tissues and cells. Used for characterizing populations of RNAs, the initial product encoded by a gene, GEPs are distinctive combinations of RNAs found in tissues and cells. Since the expression of genes is what modulates cellular function, the ability to identify a full profile of expressed RNAs is invaluable to understanding cellular function. For instance, NanoString has coupled its molecular barcode technology to a simple, reproducible hybridization protocol for high-throughput multiplex screening of transcriptomes (Geiss).
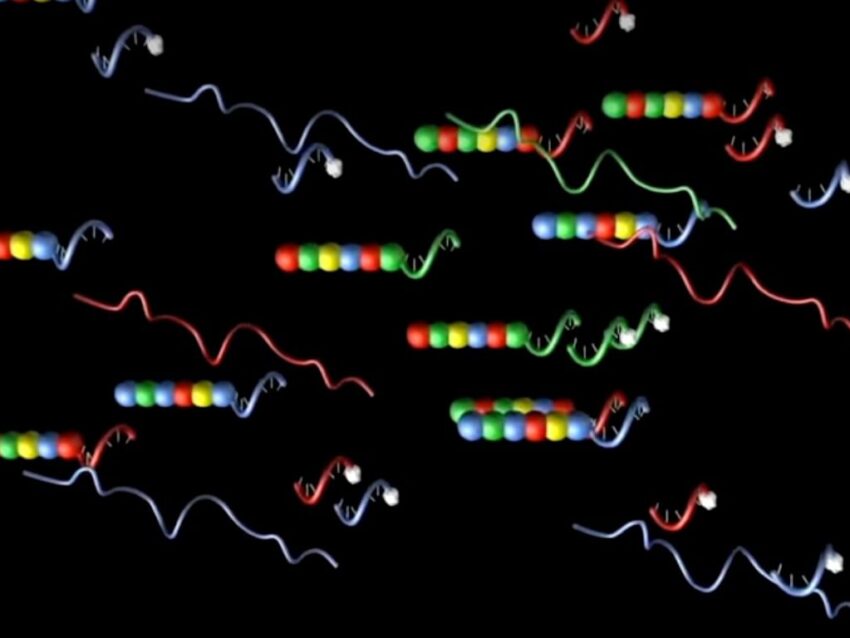
Designed in concert with specific target cDNAs to form probe panels of various gene subsets, such as for oncology or immunology applications, NanoString’s molecular barcodes incorporate RNA and DNA segments. Each RNA segment is in vitro transcribed and labeled with a specific fluorophore, which is then used to create a unique code for each gene of interest by arranging the differently colored RNA segments in a specific linear order. These fluorescent RNA barcodes are then annealed to a single-stranded DNA molecule, known as the backbone (Geiss). The reporter half of the probe pair is completed by ligating the fluorescently labeled RNA-DNA backbone to a gene-specific oligonucleotide on the 3’ end and repeated sequences on the 5’ end. A second half of the two-part probe is generated by fusing a second gene-specific sequence, taken from sequence adjacent to that of the reporter probe, to a series of 3’ repeats to form the capture probe (Geiss).

The specific color order of the labeled RNA segment in the reporter probe is used to identify the target RNA. What is innovative about this probe system is the optimized hybridization protocol that, when annealed to the target RNA, creates a tripartite structure of reporter probe, capture probe, and target molecule. After excess probe is washed away, the target RNAs are identified by the specific fluorescent signal of the bound reporter probe.
The fluorescent barcode not only identifies the gene but is also used to quantify each transcript by a direct count of each unique fluorescent signal. The direct counting of transcripts made possible by NanoString’s hybridization protocol, without the need for enzymatic or amplification steps, produces reliable data that is both unbiased and highly sensitive, even on low copy number transcripts (Geiss).
nCounter Pro’s molecular barcoding advantages
The nCounter® Pro Analysis System generates gene expression profiling data that is highly robust and reproducible. The workflow is also simple and straightforward. RNA expression profiles of 800+ genes can be analyzed from several tissue types, including FFPE tissue sections, liquid biopsies, and cell lysates. The enzyme and amplification-free probe hybridization allows for simultaneous screening of a large number of genes with sensitivity that limits the amount of tissue sample needed. Combined, the reporter and capture probes require only a 100 base pair complementary region with the target molecule for reliable gene expression measurements.

Minimizing target sequence length makes the nCounter technology highly tolerant of challenging sample types, such as FFPE, and the absence of reverse transcription and amplification steps reduces variability caused by degraded RNA and unequal copy numbers. The elegance of the fluorescent molecular barcoded probes used in the nCounter workflow eliminates several issues seen with previous gene expression analysis methods (Geiss). In RNA studies, direct hybridization of molecular barcodes can easily identify, in a highly reproducible manner, small RNAs that may be technically difficult to amplify. This reproducibility is consistent for samples across multiple users and sites, a distinct advantage over most other techniques.
Direct detection with nCounter Pro is faster than NGS and simpler than qPCR, providing single tube multiplexing for up to 800 RNA targets. With a simple protocol requiring minimal hands-on time, nCounter Pro’s assays need only crude RNA samples prepared from a variety of inputs (fresh frozen tissue, serum, plasma, PBMC and FFPE), making it great for any lab. However, it is optimized for FFPE samples, and its quick turnaround time and no library preparation makes it ideal for analyzing GEPs from clinical samples (Speranza). Furthermore, the simple data analysis available for nCounter provides publication-ready figures in just a few hours.
The elegance of the fluorescent molecular barcoded probes used in the nCounter workflow eliminates several issues seen with previous gene expression analysis methods.
An additional advantage of NanoString’s molecular barcoding technology is its adaptability. Used in bulk gene expression analysis in the nCounter Pro workflow, the original molecular barcode concept has been modified for use in two instruments that analyze a tissue’s genomics in a spatial context, GeoMx® DSP and CosMx™ SMI. The molecular barcodes follow a similar concept, with specific sequences of colored fluorescent molecules constituting the barcodes. The probes for spatial transcriptomic analysis are modified for hybridization to transcripts in tissue sections instead of in bulk tissue analysis.
The other significant difference is the barcoded probes are used in conjunction with cellular morphology markers. The morphology markers are used for visualizing regions of interest (ROI) for analysis as well as three-dimensional recreation to pinpoint localization of the targets. The barcode portion of the probe is then liberated from the ROI by UV-cleavage for quantification. Spatial and quantitative data are then integrated for analysis, and target transcripts are localized down to the subcellular level. Adapting the molecular barcode technology for use in spatial profiling is just one more way this technology can be used for advancing biology research.
References
Chen BR, Hale DC, Ciolek PJ, Runge KW. Generation and analysis of a barcode-tagged insertion mutant library in the fission yeast Schizosaccharomyces pombe. BMC Genomics. 2012;13:161. Published 2012 May 3. doi:10.1186/1471-2164-13-161
Davidsson M, Díaz-Fernández P, Torroba M, et al. Molecular barcoding of viral vectors enables mapping and optimization of mRNA trans-splicing. RNA. 2018;24(5):673-687. doi:10.1261/rna.063925.117
Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008 Mar;26(3):317-25. doi: 10.1038/nbt1385. Epub 2008 Feb 17. PMID: 18278033.
Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003 Feb 7;270(1512):313-21. doi: 10.1098/rspb.2002.2218. PMID: 12614582; PMCID: PMC1691236.
Kane MD, Jatkoe TA, Stumpf CR, Lu J, Thomas JD, Madore SJ. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 2000 Nov 15;28(22):4552-7. doi: 10.1093/nar/28.22.4552. PMID: 11071945; PMCID: PMC113865.
Li X, He Z, Zhou J. Selection of optimal oligonucleotide probes for microarrays using multiple criteria, global alignment and parameter estimation. Nucleic Acids Res. 2005 Oct 24;33(19):6114-23. doi: 10.1093/nar/gki914. PMID: 16246912; PMCID: PMC1266071.
Martel R, Shen ML, DeCorwin-Martin P, de Araujo LOF, Juncker D. Extracellular Vesicle Antibody Microarray for Multiplexed Inner and Outer Protein Analysis. ACS Sens. 2022 Dec 23;7(12):3817-3828. doi: 10.1021/acssensors.2c01750. Epub 2022 Dec 14. PMID: 36515500; PMCID: PMC9791990.
Mazurkiewicz P, Tang CM, Boone C, Holden DW. Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nat Rev Genet. 2006 Dec;7(12):929-39. doi: 10.1038/nrg1984. PMID: 17139324.
Parameswaran P, Jalili R, Tao L, et al. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 2007;35(19):e130. doi:10.1093/nar/gkm760