An Integrated Omics Approach: The Human Protein Atlas
Although humans have been using atlases for thousands of years, the word atlas was first coined in 1595 by the Flemish map maker Gerardus Mercator. In geographical terms, an atlas can be defined as a systemic collection of maps or charts that are organized with a certain structure and design. There are many kinds of atlases such as road atlases, historical atlases, and even star atlases. However, what does an atlas mean in biological terms? Traditionally in biology, atlases showed different kinds of biological data at a gross level, such as the classification of the animal kingdom or the human anatomy. Today, with the availability of complete genome sequences for humans and other organisms, atlases show information at a molecular level such as the human whole transcriptome atlas and now the Human Protein Atlas.
What is the human protein atlas?
The Human Protein Atlas represents one of the largest and most comprehensive biological databases that map the spatial distribution of all proteins in human tissues and cells at single-cell resolution. The year 2005 marked the first public release of the Human Protein Atlas project since its initiation in 2003. The main aim of the Human Protein Atlas project is to reveal the spatial distribution and expression of every human protein in different human tissues, cancer types, and cell lines, providing a functional representation of the genome that will greatly enhance our knowledge of human biology in health and disease. All the data from the Human Protein Atlas is made publicly available in the open-access knowledge resource online.
Protein profiling method for the human protein atlas
The proteome is the expression of an organism’s genome, reflecting its functional state at a given time. Proteomics is a large-scale study of proteins, expressed by a particular cell or tissue, using various biochemical and bioinformatics strategies.
Proteomics technologies that can add spatial context while analyzing protein expression are known as spatial proteomics. Spatial proteomic technologies are crucial to understanding how cells regulate protein function as they can generate detailed information such as temporal expression and protein modification including post-translational modifications and splice variants.
As proteins cannot be amplified and are easily denatured and degraded, transcriptomics is often used for the indirect measurement of protein expression.
One of the limitations of proteomics technology is that unlike transcriptomics, it is not yet possible to assess the entire repertoire of proteins from a sample. Proteins cannot be amplified and are easily denatured and degraded, resulting in sample loss and detection bias. To get around this, transcriptomics, which provides information about the location and abundance of mRNA transcripts has been used for the indirect measurement of protein expression. Transcriptomics encompasses everything relating to RNA such as the expression levels of mRNAs, their function, location, trafficking, and degradation. Hence, the Human Protein Atlas uses an integrated omics approach combining multiple technologies for mRNA and protein detection including antibody-based imaging, transcriptomics, and mass spectrometry to build a multi-dimensional spatiotemporal map of the human body.
Significant progress has been made by the Human Protein Atlas project to characterize the expression of protein-coding genes in human tissues and cells in normal and diseased states. Further, the use of proteomics and transcriptomics technologies that are compatible with archival FFPE preserved human tissues makes it possible to study human biology as directly as possible.
Antibody-based proteomics
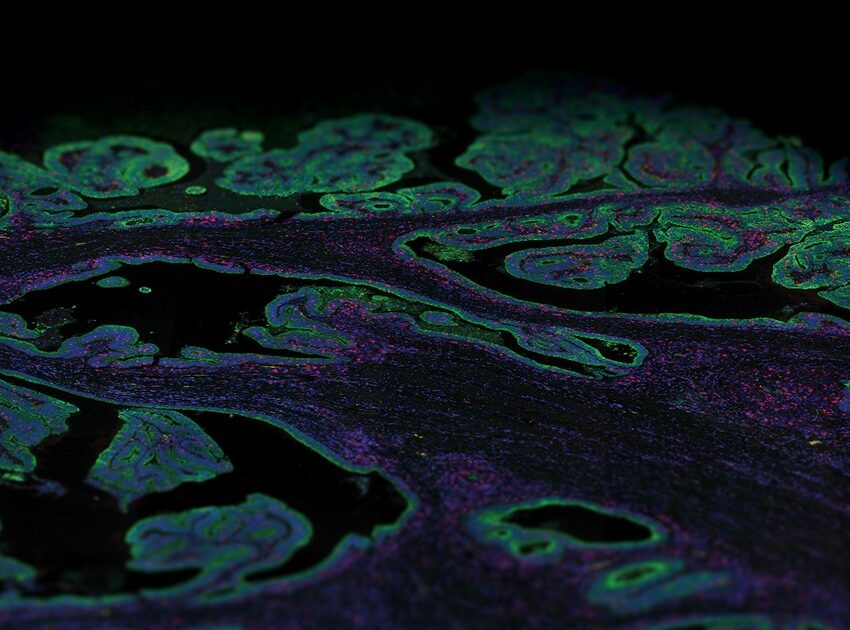
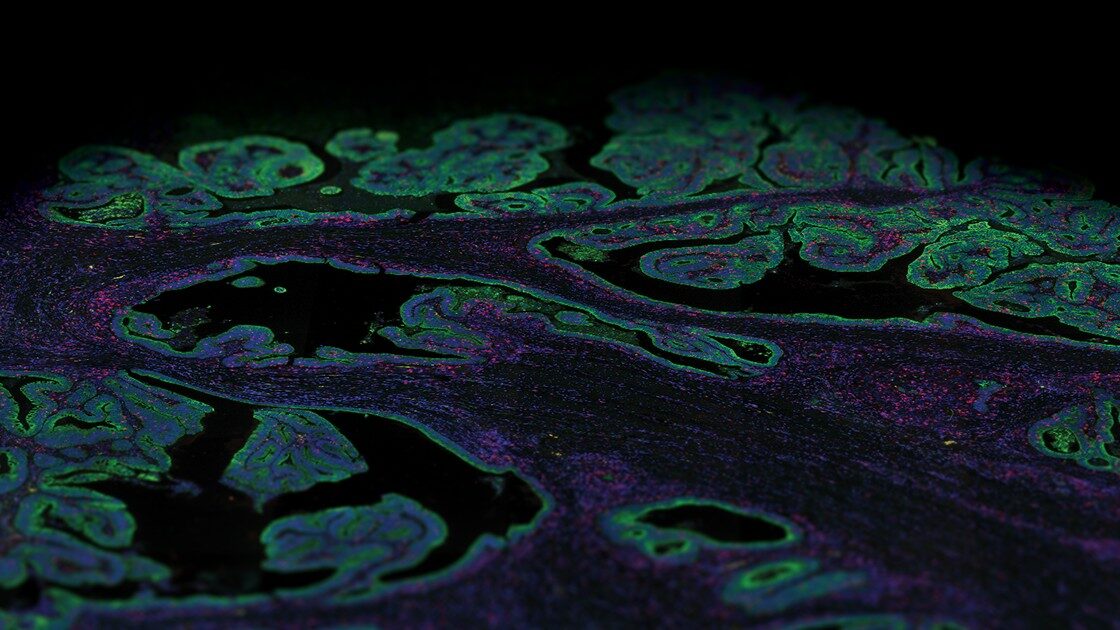
Antibody-based proteomic methods are primarily immunohistochemistry and immunofluorescence staining methods used for the detection of protein expression at the single cell or subcellular level with spatial resolution. These methods have been immensely beneficial in deciphering the phenotypic and functional architecture of tissues in health and disease.
All knowledge pages on the Human Protein Atlas provide lists of genes with direct links to corresponding high-resolution immunohistochemistry images. However, the main disadvantage of traditional immunohistochemistry is that it allows for the detection of a limited number of biomarkers per tissue section. Today, several multiplex immunohistochemical methods have emerged that differ in the way that the antibodies are tagged and use metal tags, fluorophores, DNA oligonucleotide barcodes or enzymatic detection methods that make use of mass spectrometry, fluorescence, or chromogen approaches.
As an example, NanoString‘s GeoMx® Digital Spatial Profiling (DSP) platform is a multiomics platform that allows for spatial quantification of both mRNA and protein expression within intact tissue sections The GeoMx DSP combines standard immunofluorescence techniques with oligonucleotide barcoding technology to generate expression data for tens of thousands of mRNA transcripts and 150+ proteins simultaneously with spatial resolution.
GeoMX DSP assays make use of antibody or RNA probes that are tagged with a photocleavable oligonucleotide barcode. These oligonucleotides are released upon exposure to UV light projected onto a region of interest (ROI) of the tissue. These tags are then collected and quantified using either the nCounter® Analysis System or an Illumina next-generation sequencer (NGS).
New data generated from the GeoMx DSP and CosMx SMI platforms will be valuable in gaining deep insight into aspects of the human proteome.
Another recent technology from NanoString, the CosMx™ Spatial Molecular Imager (SMI) utilizes high-plex in-situ imaging chemistry for both RNA and protein detection to provide sub-cellular or single-cell resolution. Both GeoMx DSP and CosMX SMI work with FFPE-preserved human tissues, making them ideal to study the biology of disease with biopsy samples. New data generated from the GeoMx DSP and CosMx SMI platforms will be valuable in gaining deep insight into aspects of the human proteome such as genetic variation, heterogeneity, and disease mechanisms.
Mass spectrometry-based proteomics
Mass spectrometry techniques measure the mass-to-charge ratio (m/z) of biomolecules present in a tissue sample to calculate the exact molecular weight of the sample components. Mass spectrometry has often been used in proteomics research as an alternative to antibody-based approaches and works with FFPE tissues. Mass spectrometers are composed of three basic components – an ionization source, a mass analyzer, and a detector.
Methods such as matrix-assisted laser desorption/ionization (MALDI) use laser energy to strike a matrix of biomolecules to vaporize analytes into a gas phase without fragmenting or decomposing the proteins. Secondary ion mass spectrometry (SIMS) is another technology that has a high spatial resolution down to tens of nanometers. It is used for visualizing subcellular structures but is limited to very small molecules. Mass spectrometry techniques have been applied to study various states of disease, including cancer, to discover biomarkers with diagnostic, predictive, and survival value.
Transcriptomic-based proteomics
Transcriptomic technologies are capable of a quantitative assessment of genome-wide mRNA expression from different organs, allowing for the categorization of genes based on expression levels and tissue distribution. Although transcriptomics cannot be used to study the post-translation modification of proteins and the correlation between mRNA and protein expression levels is insufficient to predict protein expression, transcriptomics nevertheless is a useful tool for the indirect measurement of protein expression.
Several technologies combine gene expression data with spatial information.
Today, several technologies combine gene expression data with spatial information. NanoString’s GeoMx DSP is built on oligonucelotide barcoding technology and can generate high throughput mRNA expression data digitally from FFPE tissue samples with resolution at the level of different tissue compartments and whole cell type populations. The GeoMx DSP can profile up to 22,000+ protein-coding genes while maintaining a wide dynamic range for the detection of low to high-expressing genes. Thus, the GeoMx DSP excels at unbiased biomarker discovery through its whole transcriptome profiling capabilities and has been instrumental in generating a spatial atlas of six different human organs: kidney, brain, colon, lymph node, liver, and pancreas.
Subdivisions of the human protein atlas
The human protein atlas is updated yearly and is one of the most accessed open-source biological databases. The current version, number 22, covers approximately 86% of the human proteome and can be subdivided into twelve different sub-atlases that are interconnected and complement each other. This enables the user to explore a protein’s tissue and organ distribution, subcellular localization, and relation to disease e.g., cancer by toggling between the different sub-atlases.
The tissue sub-atlas
The tissue sub-atlas shows the distribution of proteins across all major tissues and organs in the human body. The protein expression data for most of the tissues are derived from antibody-based protein profiling using both conventional and multiplex immunohistochemistry. Currently, the tissue sub-atlas covers 78% of protein-coding genes with expression data from 44 normal human tissue types. Expression data of mRNA is also available from 256 different normal tissue types.
The brain sub-altas
This section provides the distribution of proteins in various regions of the mammalian brain. Data is integrated from human, pig, and mouse, and each human gene is presented along with their one-to-one orthologues from pig and mouse. The gene summary pages show expression data from 13 main regions of the brain to individual nuclei and subfields for every protein-coding gene.
The single-cell type sub-atlas
This section contains expression data of protein-coding genes for single human cell types. The data is mostly derived from single-cell RNA sequencing (scRNA-seq) done on 29 human tissues and peripheral blood mononuclear cells. Genes expressed in each cell type are linked with the corresponding immunohistochemical staining of human tissues.
The tissue cell type
This section shows the expression of protein-coding genes in human cell types. Most of the data is integrated from publicly available bulk RNA-seq data. A specificity classification is used to predict which genes are enriched in each constituent cell type within an individual tissue.
The pathology sub-atlas
This section contains mRNA and protein expression data from 17 commonly occurring human cancers, along with immunohistochemical staining of tissue sections and plots showing the correlation between mRNA expression of each human protein gene and cancer patient survival.
The disease blood sub-atlas
This section shows data on protein levels in the blood of patients with different diseases and highlights proteins associated with these diseases using differential expression analysis and a disease prediction strategy based on machine learning.
The immune cell sub-atlas
This section contains single-cell information on genome-wide RNA expression profiles of human protein-coding genes covering various B- and T-cells, monocytes, granulocytes, and dendritic cells. The transcriptomic analysis covers 18 cell types isolated with cell sorting and includes classification based on specificity, distribution, and expression cluster across all immune cells.
The blood protein sub-atlas
This section describes proteins detected in human blood from mass spectrometry-based proteomics studies, published immune assay data, and a longitudinal study based on proximity extension assay. Additionally, data for genes predicted to be actively secreted to human blood is presented.
The subcellular sub-atlas
This section shows data on the spatiotemporal distribution of proteins at single-cell resolution. For each gene, the subcellular distribution of the protein by immunofluorescence and confocal microscopy in up to three different cell lines is presented. Further, the subcellular localization of the protein has been classified into one or more of 35 different organelles and fine subcellular structures.
The cell line sub-atlas
This section contains information on genome-wide RNA expression profiles of human protein-coding genes in 1,055 human cell lines, including 985 cancer cell lines. The transcriptomic analysis includes classification based on specificity analysis across 27 cancer types, distribution, and expression cluster analysis across all cell lines.
The structure sub-atlas
This section contains information about the three-dimensional structure of human proteins. All antigens with known sequences have been mapped and can be displayed on the protein structures.
The metabolic sub-atlas
This section explores the expression of protein-coding genes in the context of the human metabolic network. For proteins involved in metabolism, a metabolic summary is provided that describes the metabolic subsystems/pathways, cellular compartments, and several reactions associated with the protein. Each pathway map is accompanied by a heatmap detailing the mRNA levels across 256 different tissue types for all proteins involved in the metabolic pathway.