What is the human protein atlas used for?
In order to understand the role of each protein in various biological processes such as cell organization, signaling pathways, or disease mechanisms, it becomes essential to study protein expression with a spatial resolution.
Towards this aim, the Human Protein Atlas (HPA) project seeks to map spatial gene expression for protein-coding genes in the human body as the exact location of proteins at cellular or subcellular levels is tightly linked to protein function. The HPA provides a comprehensive overview expression of each protein categorized by organs, tissues, cell types, and disease pathology with immunohistochemically stained images of tissue samples. All the data in the Human Protein Atlas is open access and offers researchers the opportunity to explore individual genes and classes of genes and their expression profiles in the various parts of the human body.
Proteomics technologies
The HPA covers protein expression data from 15,318 genes that constitute 76% of all human protein coding genes for which there are available antibodies. Data gathered for the HPA is based on antibody-based imaging combined with transcriptomics, mass spectrometry-based proteomics, and systems biology. Antibodies reagents are commonly used as they can be implemented in a wide range of assay platforms including, multiplex and single plex Immunohistochemistry, flow cytometry, immunoprecipitation, and more. The different conditions in each application and the complexity of biological samples such as tissue-based profiling or biological fluid-based profiling ask for great functionality from the antibodies. Therefore, all antibodies produced within the Human Protein Atlas project must pass quality assurance steps before being used in any of the assays.
Tissue-based proteomics
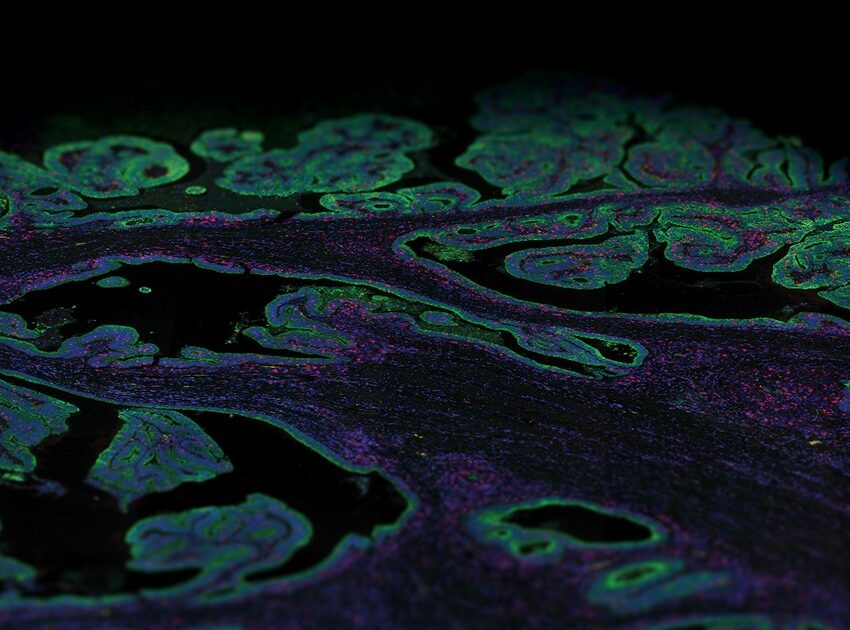
Technologies based on tissue sections are amongst the most common tools used for the detection and localization of proteins. Furthermore, many of these assays can be performed on archival tissue such as formalin-fixed paraffin-embedded (FFPE) tissue blocks, offering an opportunity for retrospective analyses for protein profiling. The Human Protein Atlas project uses a combination of spatial transcriptomics and antibody-based proteomics to create a spatiotemporal map for the protein expression at a single-cell and spatial resolution that works on FFPE tissue.
The HPA uses conventional immunohistochemistry, a microscopy-based technique, for visualizing cellular components, although it allows only a handful of markers to be visualized per tissue section, thereby limiting access to valuable information such as identifying cell variation in a homogenous population. The development of single-cell RNA sequencing (scRNA-Seq), a transcriptomics method, that allows the dissection of gene expression at single-cell resolution, has contributed to a large amount of publicly available single-cell transcriptomics datasets. The human protein atlas project integrates these multiple datasets into a single body-wide expression map of all protein-coding genes at single-cell resolution using deconvolution methods.

The correct spatial distribution of proteins is vital for their function, as often mislocalization or ectopic expression leads to diseases. Over the past decade, the advancement of several spatial technologies has enabled the detection of multiple markers from a single tissue section, capable of quantitative analysis with spatial resolution which has the potential to revolutionize data generation for the human protein atlas. Spatial multiomics technologies — which have the ability to detect both mRNA transcripts or proteins, such as NanoString’s GeoMx® Digital Spatial Profiler (DSP) and CosMx™ Spatial Molecular Imager (SMI) platforms — offer rapid, multiomic and high throughput of mRNA transcripts and over 100 protein readouts in a single experiment from a single section. Further, the profiling can be done based on morphological or phenotypical basis, adding refinement to the protein profiling methods.
Liquid biopsies
Human biological fluids such as blood and urine are routine samples for diagnostic testing and can also contain disease-associated proteins. Therefore, human biological fluids are an attractive target to profile since they are easily obtainable through noninvasive procedures.
Blood is a rich source of systemic health-related information, as physiological functions are dependent on circulating cells and proteins found in the blood.
Recently, the HPA launched a blood atlas where the proteins in the blood are described with a comprehensive analysis of all proteins predicted to be secreted from human cells (“the secretome”). The blood is a rich source of systemic health-related information as physiological functions such as the immune system, transport of nutrients and hormone regulation are all dependent on the circulating cells and proteins found in the blood. Data from the blood atlas are based on a combination of methods including flow cytometry, transcriptomics methods, mass-spectrometry, antibody-based assays, and proximity extension assays.